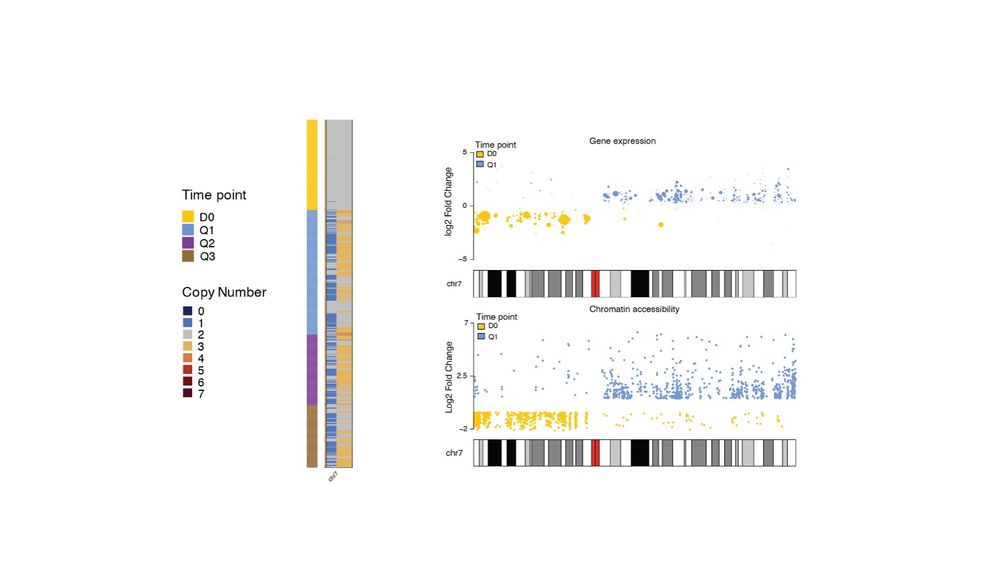
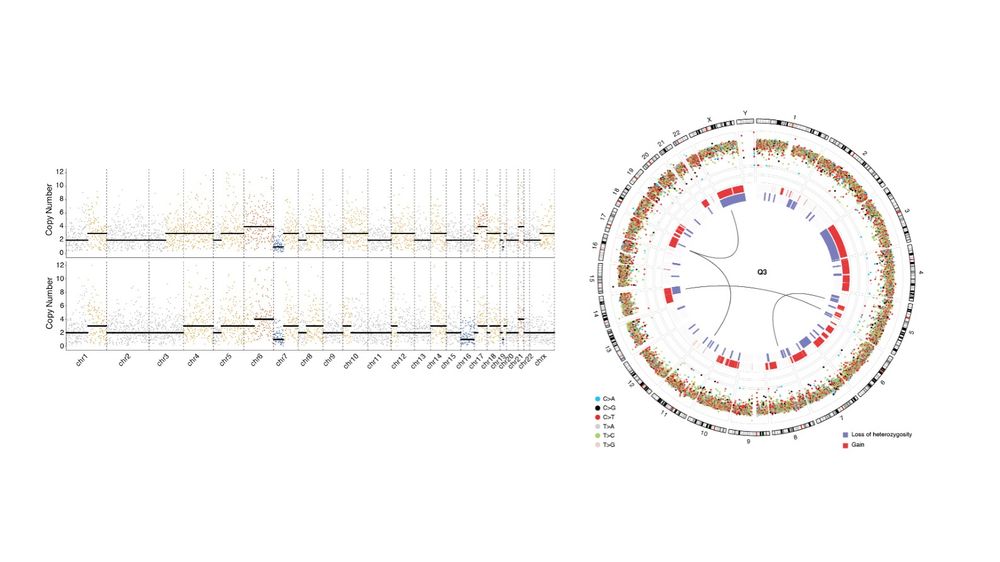
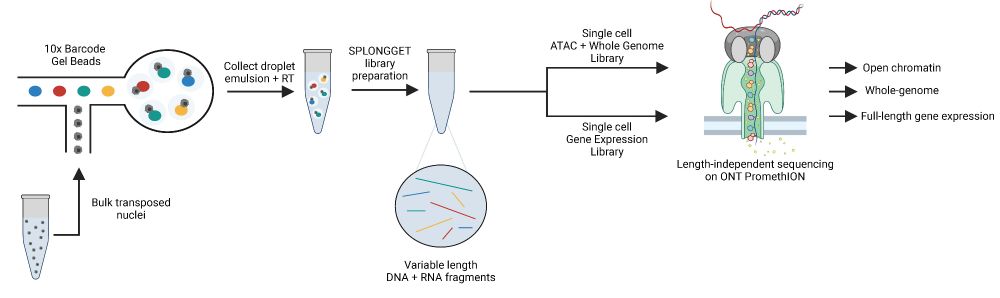
Genetics, Bioinformatics, and everything in between (she/her)
If you want to learn more, read the full pre-print here: www.biorxiv.org/content/10.1....

If you want to learn more, read the full pre-print here: www.biorxiv.org/content/10.1....