Alexander Belyy
@alexanderbelyy.bsky.social
Assistant professor, University of Groningen, NL.
Studying bacterial toxins and actin cytoskeleton using cryo-EM.
Studying bacterial toxins and actin cytoskeleton using cryo-EM.
We bought a nail dryer from Amazon for 20 euro 🙂
August 14, 2025 at 8:20 AM
We bought a nail dryer from Amazon for 20 euro 🙂
Great work by @jakecolautti.bsky.social @dshatskiy.bsky.social Eileen Bates, Nathan P. Bullen & John C. Whitney
May 1, 2025 at 3:37 PM
Great work by @jakecolautti.bsky.social @dshatskiy.bsky.social Eileen Bates, Nathan P. Bullen & John C. Whitney
Many thanks to @dshatskiy.bsky.social, Athul Sivan, and Roland Wedlich-Söldner for the excellent work!
#Actin #Ftractin #Cytoskeleton #Microscopy #StructuralBiology #CryoEM #Science
#Actin #Ftractin #Cytoskeleton #Microscopy #StructuralBiology #CryoEM #Science
February 11, 2025 at 5:38 PM
Many thanks to @dshatskiy.bsky.social, Athul Sivan, and Roland Wedlich-Söldner for the excellent work!
#Actin #Ftractin #Cytoskeleton #Microscopy #StructuralBiology #CryoEM #Science
#Actin #Ftractin #Cytoskeleton #Microscopy #StructuralBiology #CryoEM #Science
Finally, based on the structural data, we designed an optimized version of F-tractin. The result? Great labeling and minimal disturbance to actin dynamics! 🚀
February 11, 2025 at 5:38 PM
Finally, based on the structural data, we designed an optimized version of F-tractin. The result? Great labeling and minimal disturbance to actin dynamics! 🚀
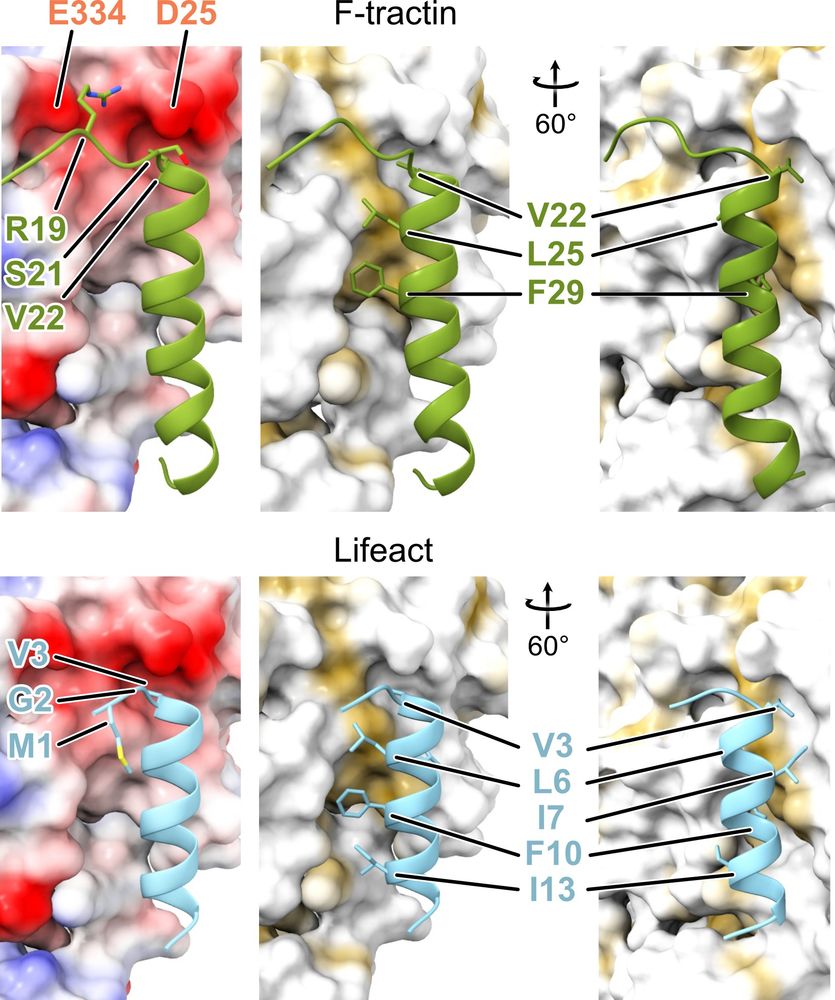
These structural results and the biochemical data show that the previously reported differences between these peptides originate from the flexible areas of F-tractin or from differential expression levels, rather than from fundamentally different modes of interaction with F-actin.
February 11, 2025 at 5:38 PM
These structural results and the biochemical data show that the previously reported differences between these peptides originate from the flexible areas of F-tractin or from differential expression levels, rather than from fundamentally different modes of interaction with F-actin.
Unexpectedly, this binding site is nearly identical to the one of #Lifeact - the most popular probe for visualization of actin in living cells!
February 11, 2025 at 5:38 PM
Unexpectedly, this binding site is nearly identical to the one of #Lifeact - the most popular probe for visualization of actin in living cells!
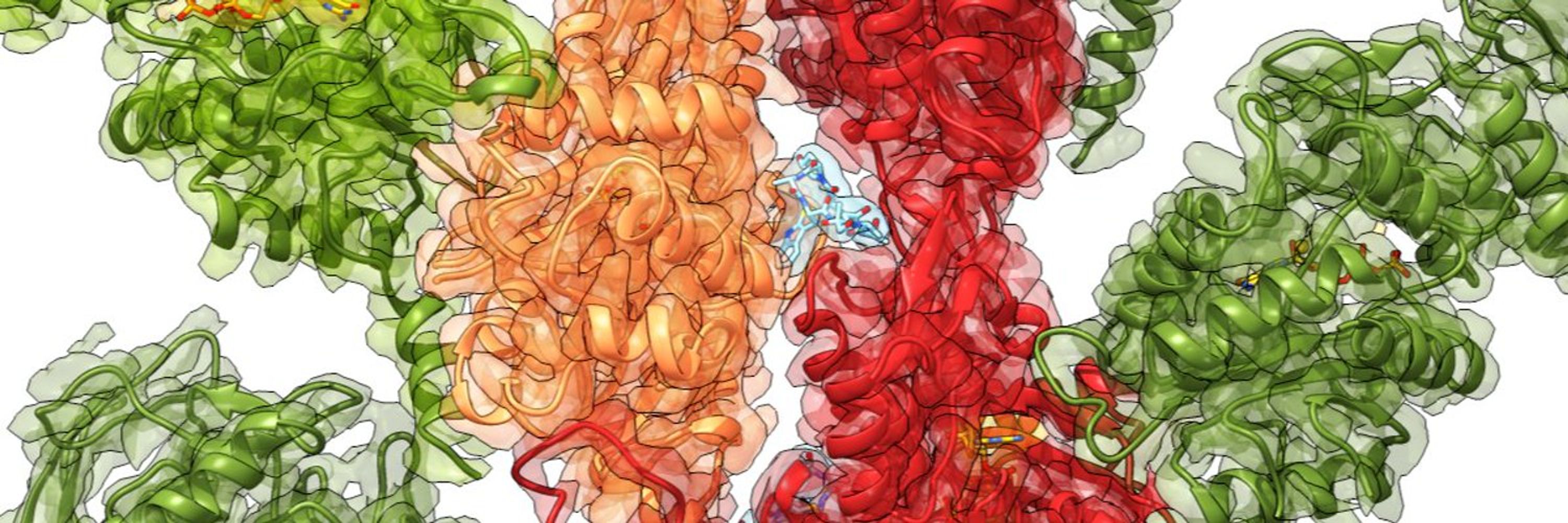
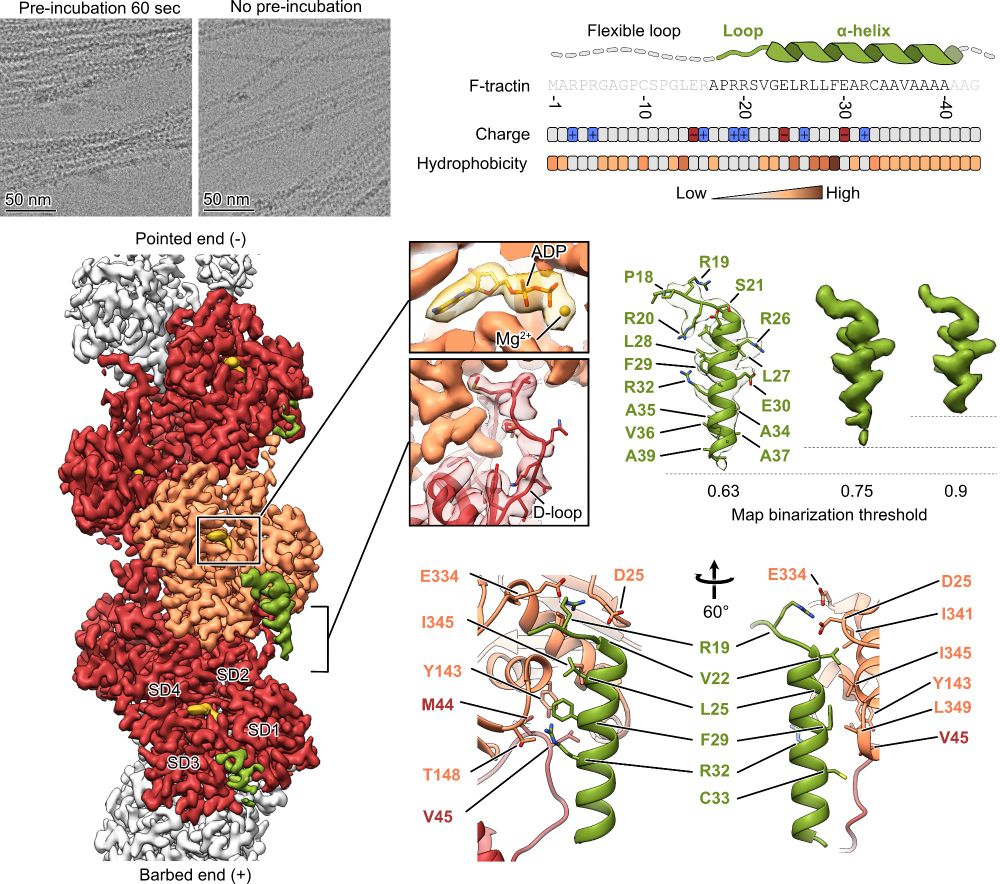
Using cryo-EM 🔬, we determined the structure of the F-tractin–F-actin complex at high resolution. We found its specific binding site, which explained its actin-binding properties!
Check out these structural insights! 📸
Check out these structural insights! 📸
February 11, 2025 at 5:38 PM
Using cryo-EM 🔬, we determined the structure of the F-tractin–F-actin complex at high resolution. We found its specific binding site, which explained its actin-binding properties!
Check out these structural insights! 📸
Check out these structural insights! 📸
F-tractin is a small peptide derived from the rat ITPKA protein that binds F-actin. It’s a go-to tool for actin #cytoskeleton visualization in live cells. But… how does it work at the molecular level? 🤔🔍
February 11, 2025 at 5:38 PM
F-tractin is a small peptide derived from the rat ITPKA protein that binds F-actin. It’s a go-to tool for actin #cytoskeleton visualization in live cells. But… how does it work at the molecular level? 🤔🔍

