
Prev: @jura.bsky.social, PhD with Debora Marks
Website: alannawzadamin.github.io
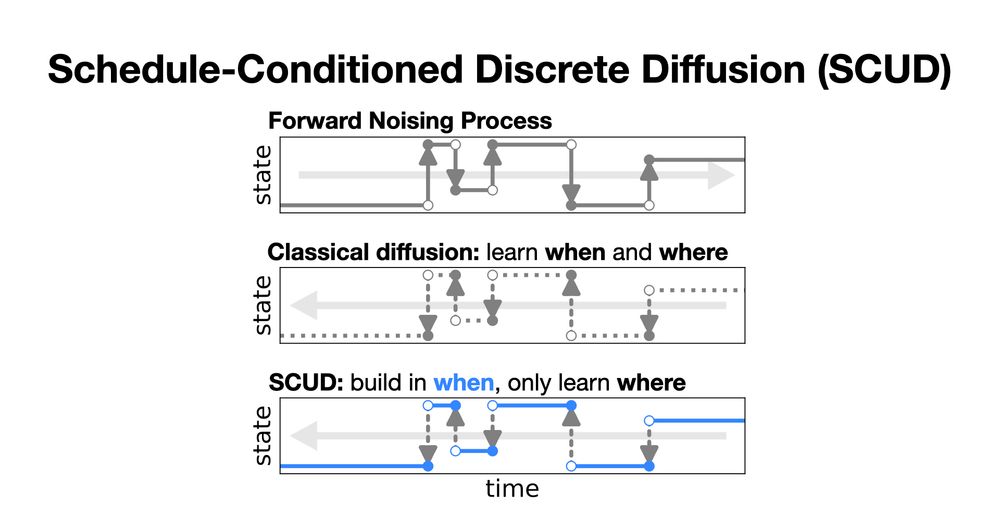
More experiments and details of our linear algebra in the paper! Come say hi at ICML! 7/7
Paper: arxiv.org/abs/2506.19598
Code: github.com/AlanNawzadAm...

More experiments and details of our linear algebra in the paper! Come say hi at ICML! 7/7
Paper: arxiv.org/abs/2506.19598
Code: github.com/AlanNawzadAm...





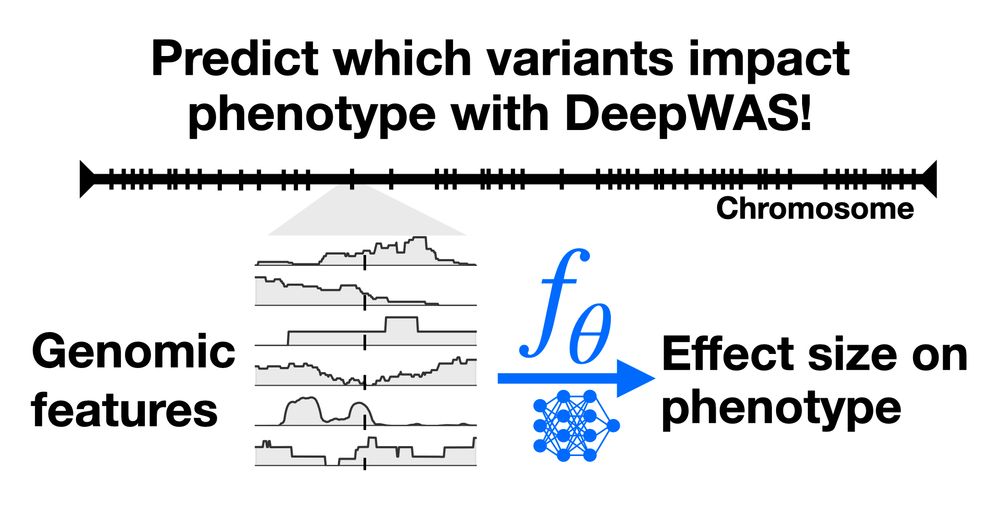









SoTA methods search the space of sequences by iteratively suggesting mutations. But the space of antibodies is huge! CloneBO builds a prior on mutations that make strong and stable binders in our body to optimize antibodies in silico. 2/7

SoTA methods search the space of sequences by iteratively suggesting mutations. But the space of antibodies is huge! CloneBO builds a prior on mutations that make strong and stable binders in our body to optimize antibodies in silico. 2/7
w/ Nat Gruver, Yilun Kuang, Lily Li, @andrewgwils.bsky.social and the team at Big Hat! 1/7
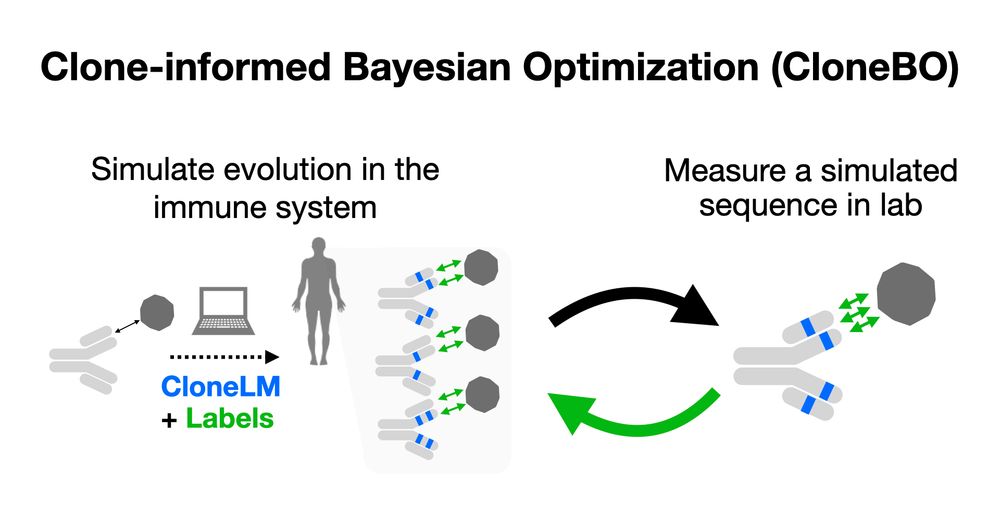
w/ Nat Gruver, Yilun Kuang, Lily Li, @andrewgwils.bsky.social and the team at Big Hat! 1/7


