Stay tuned for more on this!
Stay tuned for more on this!
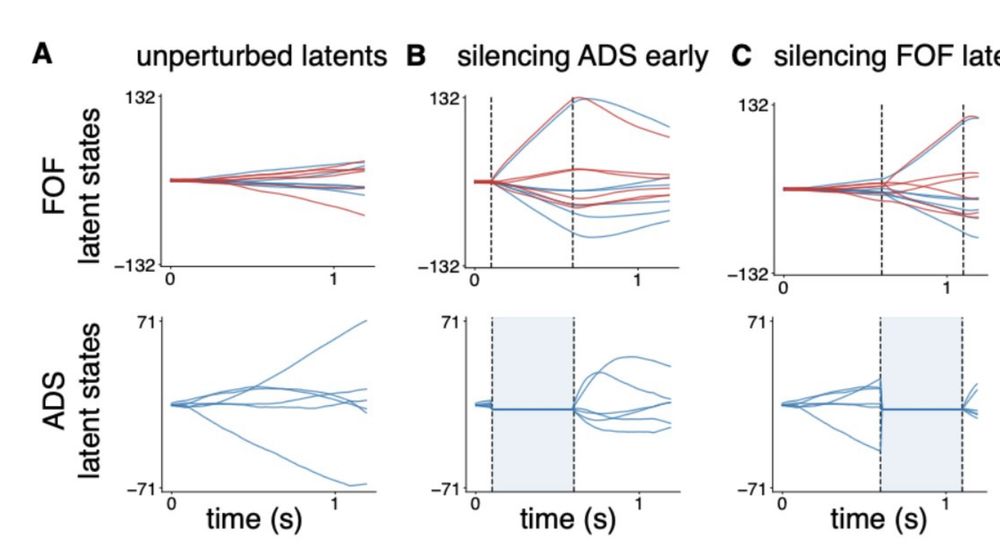
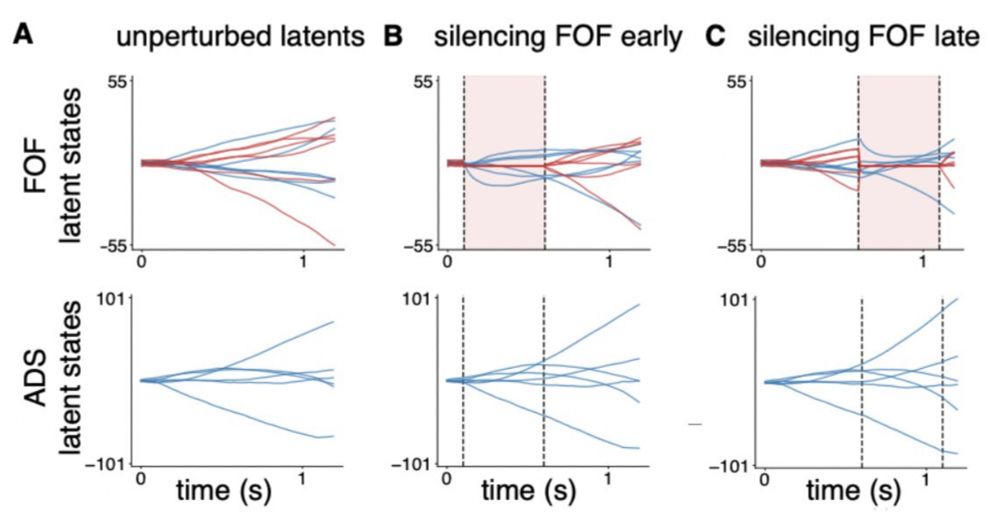
(Note that we did not fit the model on perturbed data.)
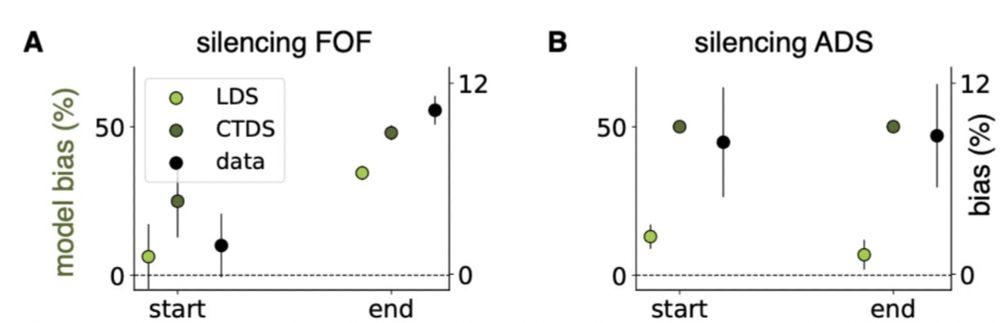
(Note that we did not fit the model on perturbed data.)
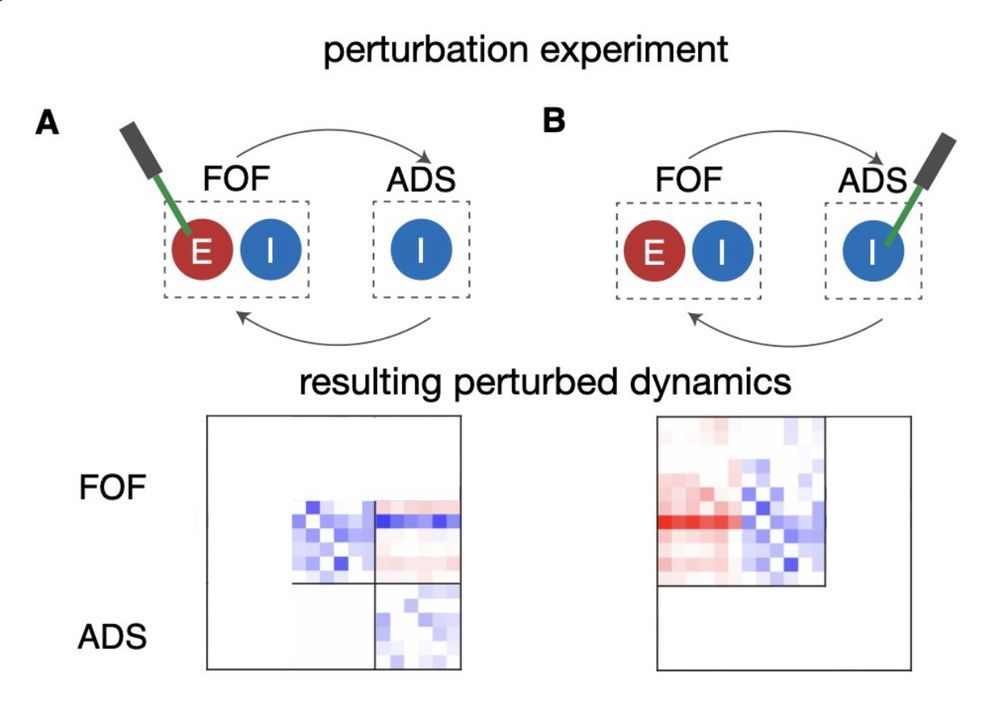
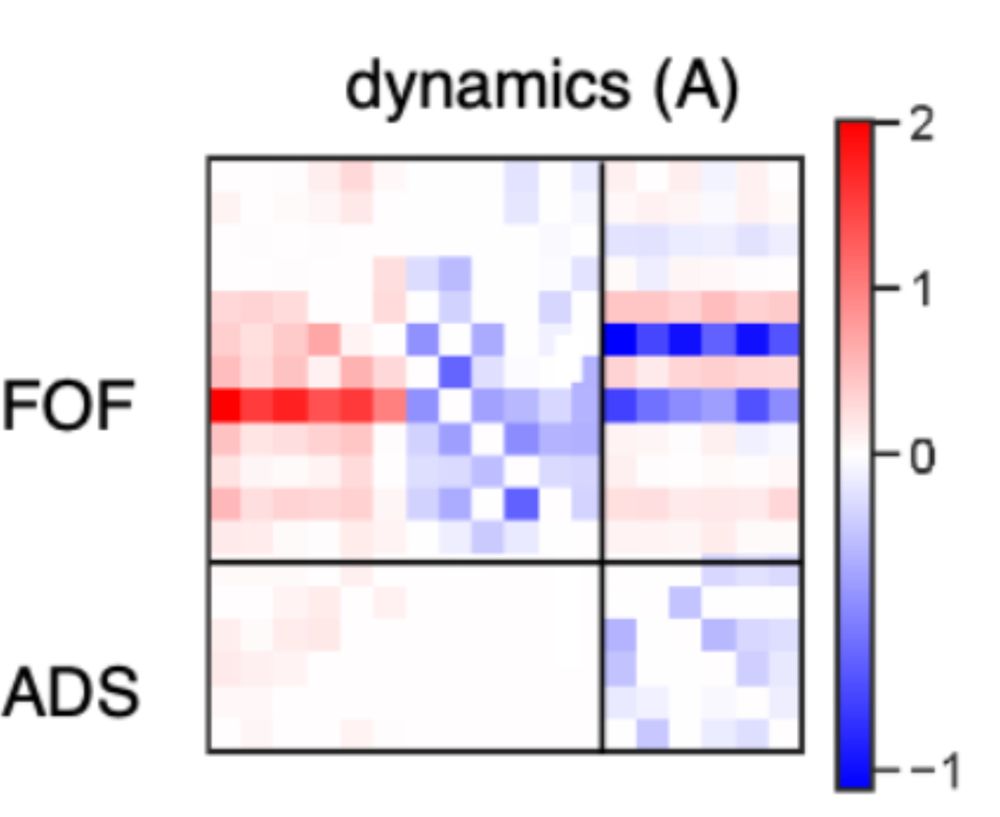
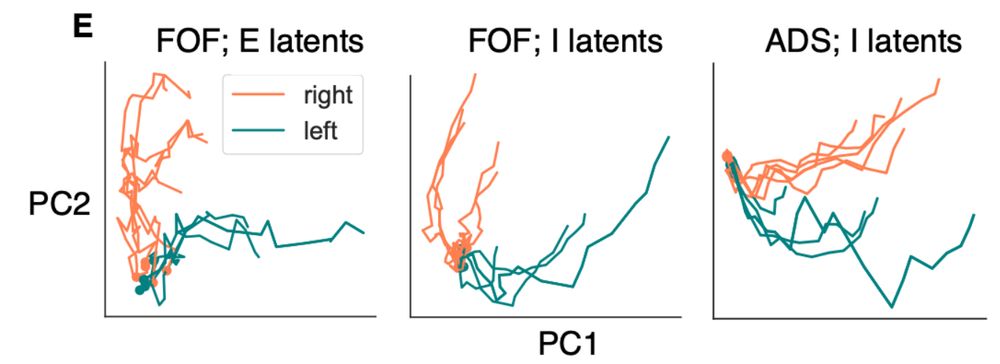
We also extend it to multi-region settings!
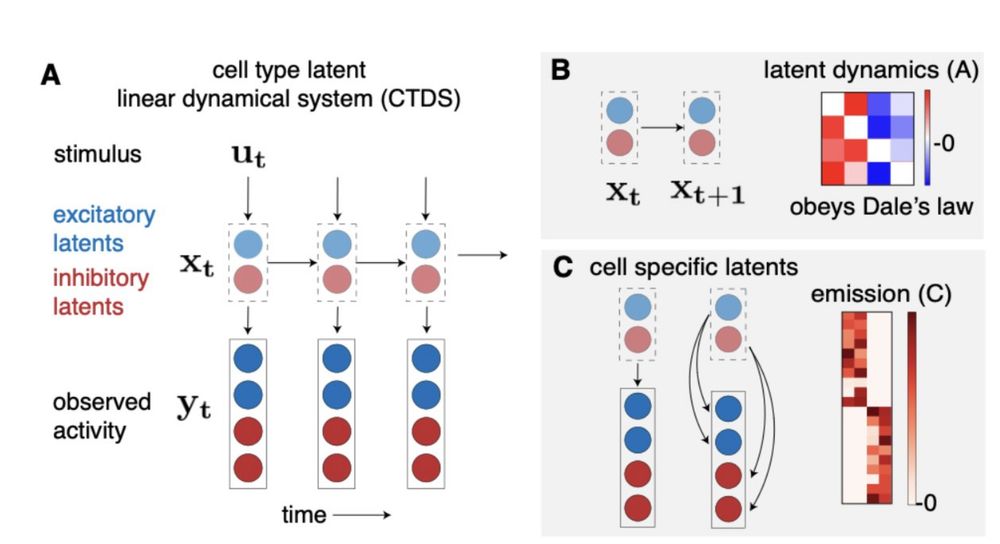
We also extend it to multi-region settings!

