Adam Cribbs
@adamcribbs.bsky.social
#MRC CDA fellow,
University of Oxford, long-read sequencing| single-cell technology development| multiple myeloma| bone cancer
University of Oxford, long-read sequencing| single-cell technology development| multiple myeloma| bone cancer
📢 New preprint alert!
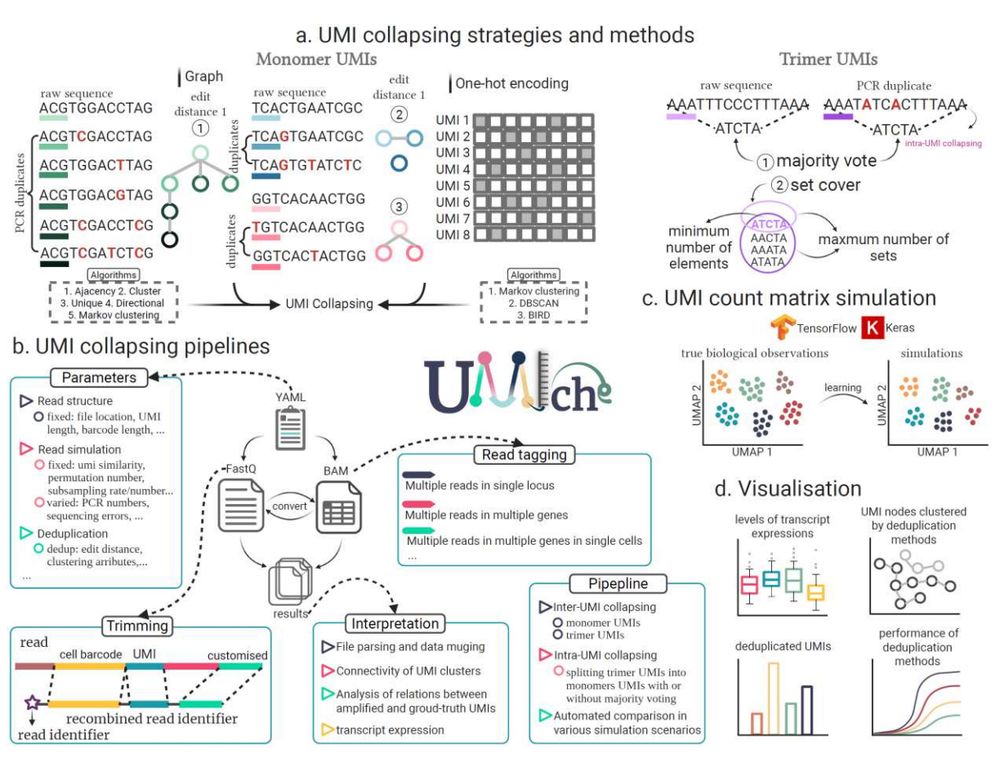
We present UMIche, a modular, open-source platform to improve UMI-based molecular quantification in bulk and scRNA-seq data.
→ Integrated pipelines
→ Monomer & homotrimer UMIs
→ Deep learning-based simulation
🔗 doi.org/10.21203/rs....
We present UMIche, a modular, open-source platform to improve UMI-based molecular quantification in bulk and scRNA-seq data.
→ Integrated pipelines
→ Monomer & homotrimer UMIs
→ Deep learning-based simulation
🔗 doi.org/10.21203/rs....
May 6, 2025 at 9:19 AM
📢 New preprint alert!
We present UMIche, a modular, open-source platform to improve UMI-based molecular quantification in bulk and scRNA-seq data.
→ Integrated pipelines
→ Monomer & homotrimer UMIs
→ Deep learning-based simulation
🔗 doi.org/10.21203/rs....
We present UMIche, a modular, open-source platform to improve UMI-based molecular quantification in bulk and scRNA-seq data.
→ Integrated pipelines
→ Monomer & homotrimer UMIs
→ Deep learning-based simulation
🔗 doi.org/10.21203/rs....
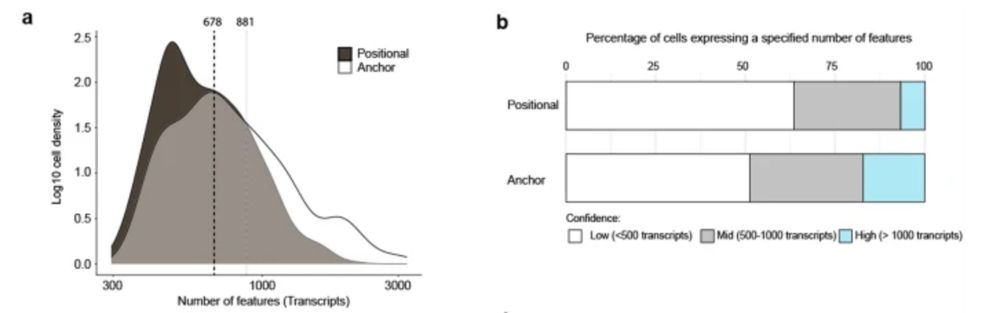
The effects of adding an anchor sequence are a significantly increased number of features per cell over and above not having an anchor (Positional).
February 16, 2025 at 10:47 AM
The effects of adding an anchor sequence are a significantly increased number of features per cell over and above not having an anchor (Positional).
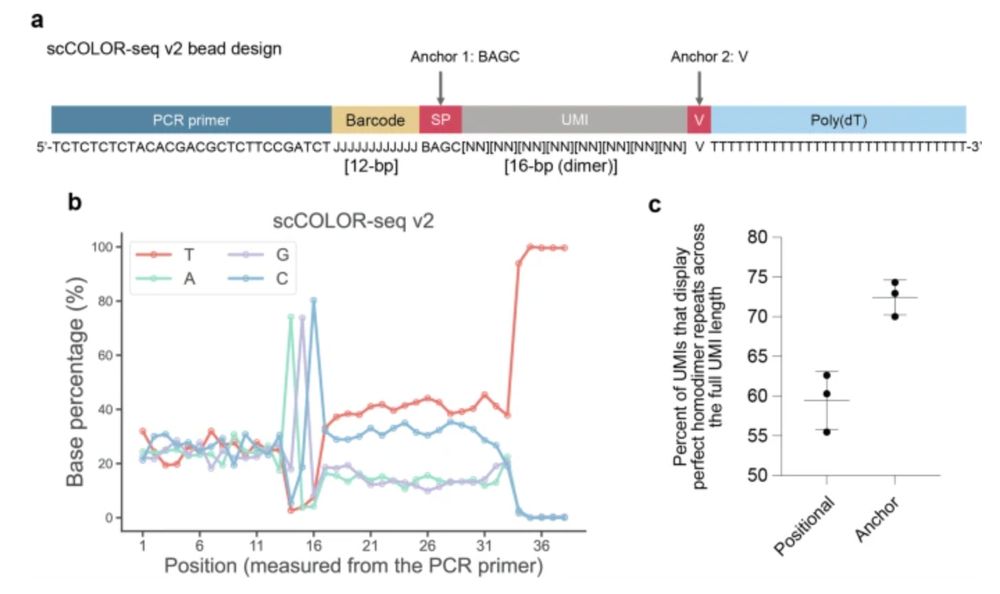
By integrating an anchor sequence before and after the UMI, we observed a significant reduction in technical variability, an increase in UMI accuracy, leading to more accurate gene expression measurements.
February 16, 2025 at 10:47 AM
By integrating an anchor sequence before and after the UMI, we observed a significant reduction in technical variability, an increase in UMI accuracy, leading to more accurate gene expression measurements.
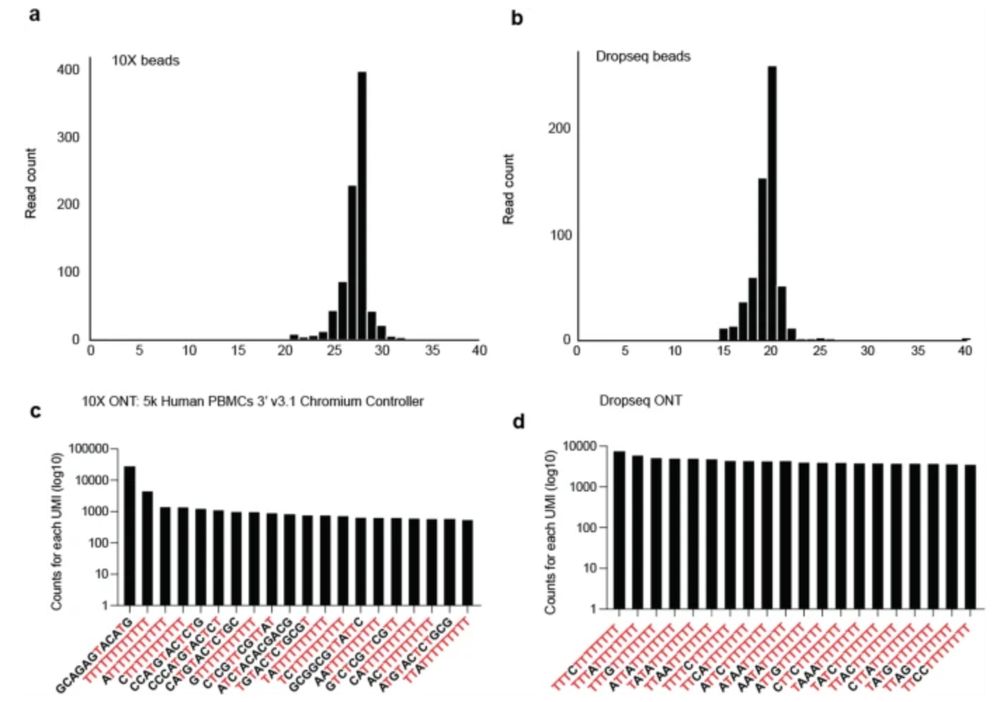
Turns out for droplet based single-cell methods, beads are truncated and this leads to several issues with downstream artefacts. Mainly, there is an overrepresentation of Ts in the UMI that result from truncated bead synthesis.
February 16, 2025 at 10:47 AM
Turns out for droplet based single-cell methods, beads are truncated and this leads to several issues with downstream artefacts. Mainly, there is an overrepresentation of Ts in the UMI that result from truncated bead synthesis.

