
Austin Daigle
@adaigle.bsky.social
PhD Candidate in Bioinformatics & Computational Biology at UNC, coadvised by Dan Schrider & Parul Johri. I work on population genetics, transposon detection, and simulation-based inference—simulating evolution because real evolution takes way too long.
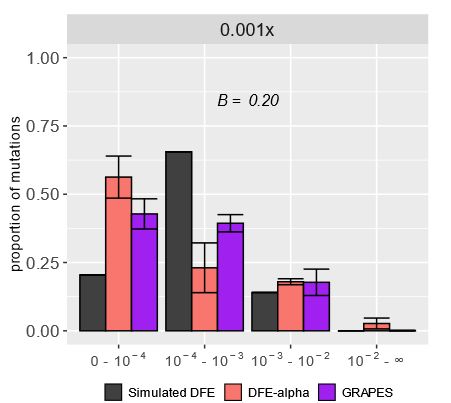
If HRI was the cause mis-inference, we’d expect it to be caused by the reduced levels of effective recombination in selfers. Indeed, when we ran simulations with lower recombination (but no selfing), we saw the same patterns of DFE mis-inference.
March 5, 2025 at 6:36 PM
If HRI was the cause mis-inference, we’d expect it to be caused by the reduced levels of effective recombination in selfers. Indeed, when we ran simulations with lower recombination (but no selfing), we saw the same patterns of DFE mis-inference.
We hypothesized this mis-inference was caused by HRI, where linked deleterious mutations interact and reduce the efficacy of selection. The site frequency spectrum (SFS) had a U-shape at high selfing rates, a pattern often linked to HRI and not modeled by current DFE inference approaches.

March 5, 2025 at 6:36 PM
We hypothesized this mis-inference was caused by HRI, where linked deleterious mutations interact and reduce the efficacy of selection. The site frequency spectrum (SFS) had a U-shape at high selfing rates, a pattern often linked to HRI and not modeled by current DFE inference approaches.
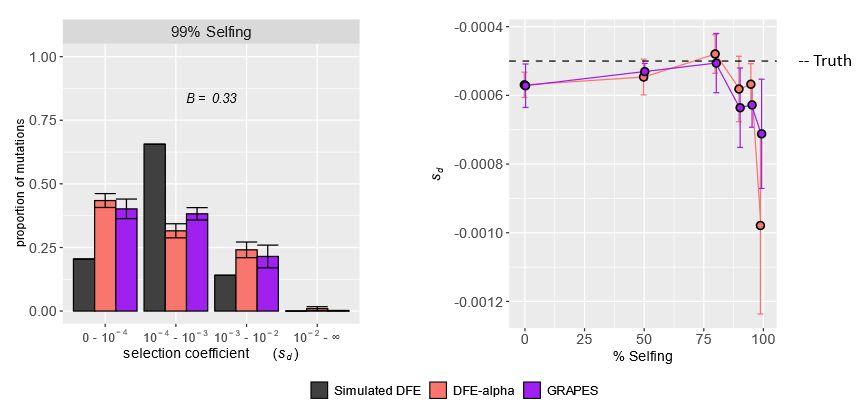
In simulated highly selfing populations, the DFE was mis-inferred by two unique DFE inference methods—nearly neutral and strongly deleterious mutations were overestimated, while mildly deleterious ones were underestimated.
March 5, 2025 at 6:36 PM
In simulated highly selfing populations, the DFE was mis-inferred by two unique DFE inference methods—nearly neutral and strongly deleterious mutations were overestimated, while mildly deleterious ones were underestimated.
Excited to share my first PhD project with my mentor, @johriparul.bsky.social! We examine how Hill–Robertson interference (HRI) in highly selfing species biases estimates of the distribution of fitness effects of new mutations (DFE).
doi.org/10.1093/evol...
@journal-evo.bsky.social #popgen #evobio
doi.org/10.1093/evol...
@journal-evo.bsky.social #popgen #evobio

March 5, 2025 at 6:36 PM
Excited to share my first PhD project with my mentor, @johriparul.bsky.social! We examine how Hill–Robertson interference (HRI) in highly selfing species biases estimates of the distribution of fitness effects of new mutations (DFE).
doi.org/10.1093/evol...
@journal-evo.bsky.social #popgen #evobio
doi.org/10.1093/evol...
@journal-evo.bsky.social #popgen #evobio

