Special thanks to our core contributors:
@bkmi.bsky.social, Bing Yan, Xiang Fu, Guang-Horng Liu (and of course Carles Domingo-Enrich).
Special thanks to our core contributors:
@bkmi.bsky.social, Bing Yan, Xiang Fu, Guang-Horng Liu (and of course Carles Domingo-Enrich).
The benchmark features real, drug-like molecules from the SPICE dataset, and we hope it drives direct and tangible progress in sampling for computational chemistry (coming soon).

The benchmark features real, drug-like molecules from the SPICE dataset, and we hope it drives direct and tangible progress in sampling for computational chemistry (coming soon).
By exploiting a factorization of the optimal transition density (a Schrödinger bridge), our new loss enables heavy reuse of simulations and energy evaluations.
By exploiting a factorization of the optimal transition density (a Schrödinger bridge), our new loss enables heavy reuse of simulations and energy evaluations.
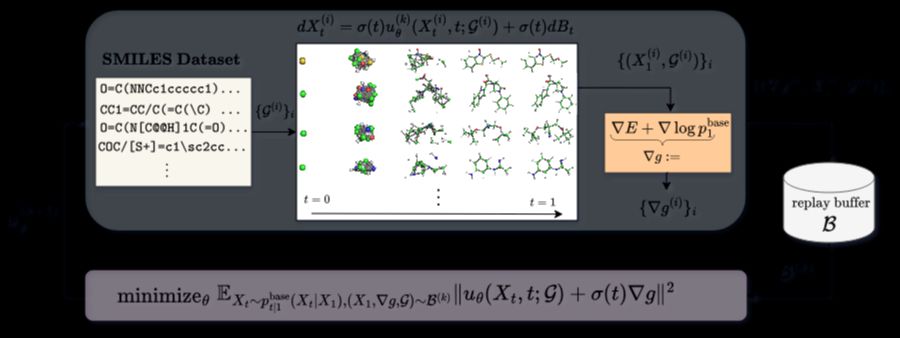
By exploiting a factorization of the optimal transition density (a Schrödinger bridge), our new loss enables heavy reuse of simulations and energy evaluations.

By exploiting a factorization of the optimal transition density (a Schrödinger bridge), our new loss enables heavy reuse of simulations and energy evaluations.

