"Ensemble docking for intrinsically disordered proteins"
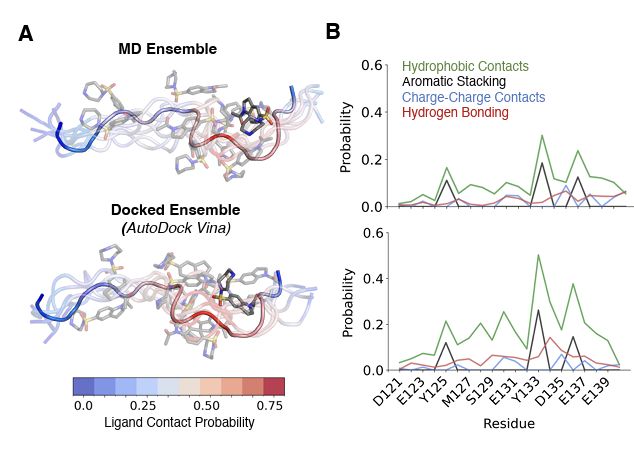
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...


