at Norwegian University of Life Sciences
Read the paper by @siribirkeland.bsky.social and @trhvidsten.bsky.social with contributions from several UPSC researchers.
By comparing dozens of genomes, they offer fresh insights into the genetic foundations of woodiness and perenniality in angiosperms.
Read the paper by @siribirkeland.bsky.social and @trhvidsten.bsky.social with contributions from several UPSC researchers.
By comparing dozens of genomes, they offer fresh insights into the genetic foundations of woodiness and perenniality in angiosperms.
buff.ly/yXVfgnP
#PlantaePSRW
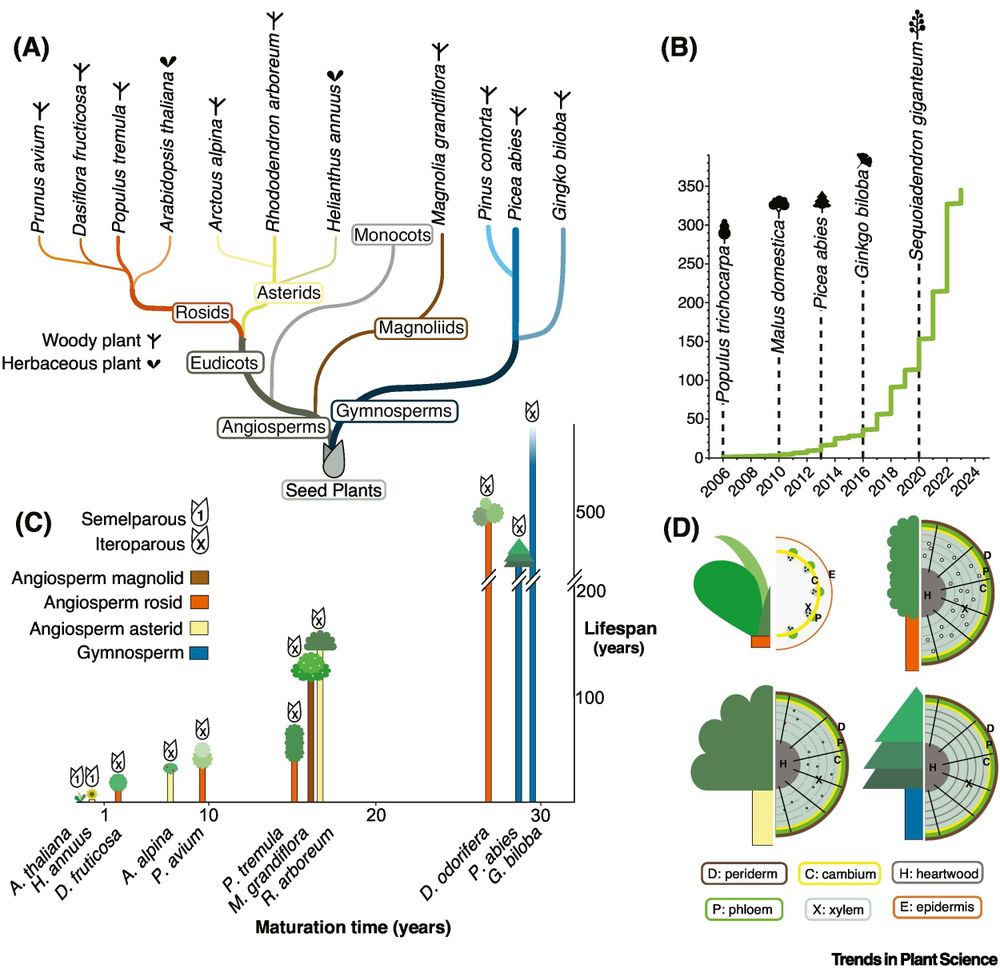
buff.ly/yXVfgnP
#PlantaePSRW
#plantscience

#plantscience
All our PhD candidates together for the first time – a week of project presentations, hands-on workshops, industry talks, and an amazing sailing social on the Med. Science + collaboration + tapas = success!
#HoloGen #PhDLife #Microbiome #MSCADN

All our PhD candidates together for the first time – a week of project presentations, hands-on workshops, industry talks, and an amazing sailing social on the Med. Science + collaboration + tapas = success!
#HoloGen #PhDLife #Microbiome #MSCADN
Try the app out here👇: lauragarciaromanach.shinyapps.io/popul_r_mini/

Try the app out here👇: lauragarciaromanach.shinyapps.io/popul_r_mini/
By comparing trees grown outdoors and in lab environments, they reveal seasonal genetic patterns and highlight both the strengths and limitations of lab-based experiments to study seasonality.
👉Read more here: www.upsc.se/about-upsc/n...

By comparing trees grown outdoors and in lab environments, they reveal seasonal genetic patterns and highlight both the strengths and limitations of lab-based experiments to study seasonality.
👉Read more here: www.upsc.se/about-upsc/n...

www.biorxiv.org/content/10.1...
We've leveraged methods developed for multi-omics to investigate a holo-omic system (🐷🦠), and predicted host-microbiome interactions between important features for observed cross-omic data patterns 🔍📊
www.biorxiv.org/content/10.1...
We've leveraged methods developed for multi-omics to investigate a holo-omic system (🐷🦠), and predicted host-microbiome interactions between important features for observed cross-omic data patterns 🔍📊
doi.org/10.1038/d415...

doi.org/10.1038/d415...
The positions are part of @hologen.bsky.social
www.jobbnorge.no/en/available...
Deadline: January 17th. @microbesrule.bsky.social @sabinallarosa.bsky.social

The positions are part of @hologen.bsky.social
www.jobbnorge.no/en/available...
Deadline: January 17th. @microbesrule.bsky.social @sabinallarosa.bsky.social
1/4
1/4
We analyzed 470 (!) angiosperm genomes and identified and dated 132 whole-genome duplication events. Importantly, we observed that WGD occurrence dates are not randomly distributed, but clustered at times of major environmental upheaval.
We analyzed 470 (!) angiosperm genomes and identified and dated 132 whole-genome duplication events. Importantly, we observed that WGD occurrence dates are not randomly distributed, but clustered at times of major environmental upheaval.


