I’m combining biophysical modeling with machine learning to study genome folding.
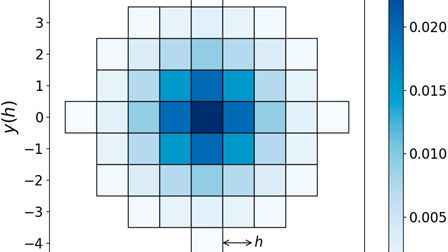
We built dLEM - a differentiable Loop Extrusion Model that bridges biophysics and machine learning for 3D genome folding.
dLEM makes loop extrusion learnable and interpretable—predicting how genomes fold and how they respond to perturbations.
🧵
We built dLEM - a differentiable Loop Extrusion Model that bridges biophysics and machine learning for 3D genome folding.
dLEM makes loop extrusion learnable and interpretable—predicting how genomes fold and how they respond to perturbations.
🧵