
Haven't read it fully, yet! Sounds interesting...

Haven't read it fully, yet! Sounds interesting...
🎉🎉🎉 I have been selected as the Indian Ambassador for @biophysicalsoc.bsky.social seventh cohort (2026-2028)!!! Eagerly looking forward to working with other ambassadors to promote biophysics and bioinfo in India. 🥳 🙌
biophysics.cld.bz/Biophysical-...

🎉🎉🎉 I have been selected as the Indian Ambassador for @biophysicalsoc.bsky.social seventh cohort (2026-2028)!!! Eagerly looking forward to working with other ambassadors to promote biophysics and bioinfo in India. 🥳 🙌
biophysics.cld.bz/Biophysical-...
Deadline: 16 January 2026
🔗 eccb2026.org/call-tutoria...

Deadline: 16 January 2026
🔗 eccb2026.org/call-tutoria...

www.madebyolof.com/bluesky-wrap...

www.madebyolof.com/bluesky-wrap...
www.cell.com/biophysj/ful...
@florencetama.bsky.social @frettinglab.bsky.social @biophysicalsoc.bsky.social @biophysj.bsky.social

www.cell.com/biophysj/ful...
@florencetama.bsky.social @frettinglab.bsky.social @biophysicalsoc.bsky.social @biophysj.bsky.social
This work shows how structural systems biology can:
✔️ Improve phenotype prediction
✔️ Enable rational strain design
✔️ Bridge molecular mechanism ↔ metabolic engineering
📂 Code & data: github.com/raghuyennama...
7/7

This work shows how structural systems biology can:
✔️ Improve phenotype prediction
✔️ Enable rational strain design
✔️ Bridge molecular mechanism ↔ metabolic engineering
📂 Code & data: github.com/raghuyennama...
7/7
Using Foldseek, we clustered ~4,200 AlphaFold structures:
• 563 fold clusters
• Levan enzymes grouped into 4 structural clusters
• Enabled functional hints for 7 uncharacterized proteins 6/7
Using Foldseek, we clustered ~4,200 AlphaFold structures:
• 563 fold clusters
• Levan enzymes grouped into 4 structural clusters
• Enabled functional hints for 7 uncharacterized proteins 6/7
We built a Boolean gene regulatory network (BooleSim) for levan biosynthesis.
➡️ Identified stable core genes (e.g., sacB, sacY, levB)
➡️ Captured oscillatory regulators (abrB, gmuR, gmuE) across conditions. 5/7
We built a Boolean gene regulatory network (BooleSim) for levan biosynthesis.
➡️ Identified stable core genes (e.g., sacB, sacY, levB)
➡️ Captured oscillatory regulators (abrB, gmuR, gmuE) across conditions. 5/7
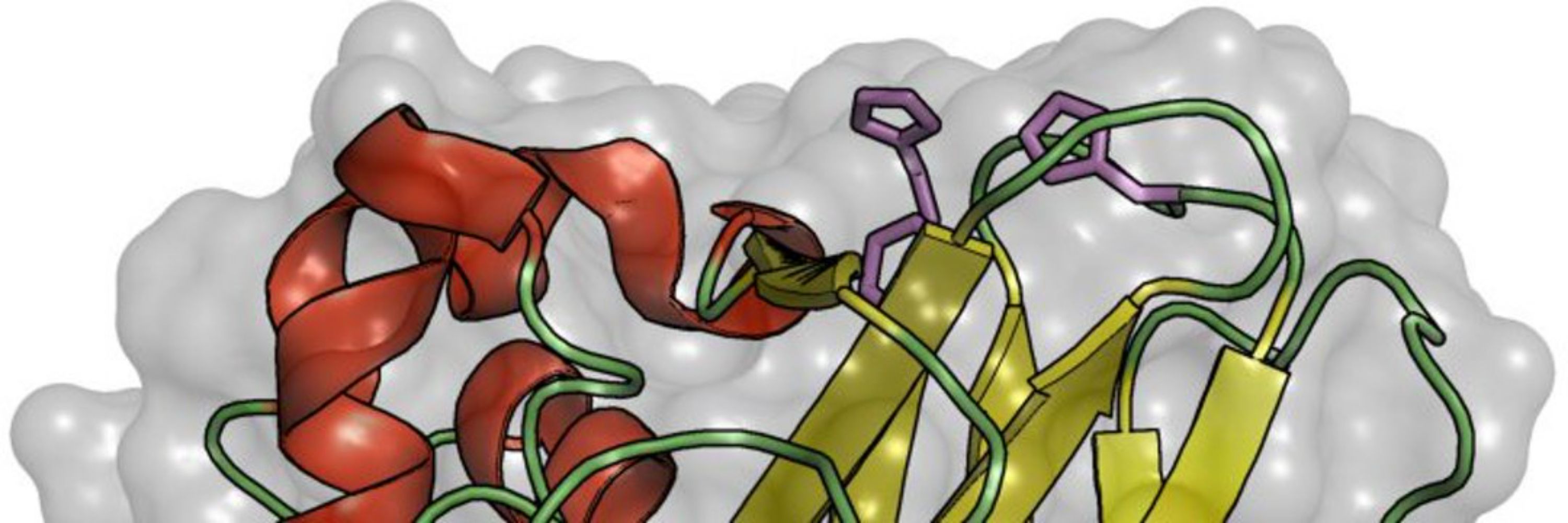
Levansucrase has many structures, many mutants, many catalytic states.
By embedding these into the pathway, we connect polymer length, mechanism, and metabolic context — not just fluxes. 4/7
Levansucrase has many structures, many mutants, many catalytic states.
By embedding these into the pathway, we connect polymer length, mechanism, and metabolic context — not just fluxes. 4/7
A structure-annotated GEM where:
• 508 PDB structures
• 168 proteins
• 331 reactions (~27% of the model) are explicitly linked. This is NOT a cosmetic improvement. The model has AF2 structures as well. 3/7
A structure-annotated GEM where:
• 508 PDB structures
• 168 proteins
• 331 reactions (~27% of the model) are explicitly linked. This is NOT a cosmetic improvement. The model has AF2 structures as well. 3/7

