doi.org/10.1038/s415...
It was a long journey – 16 months from initial submission to acceptance. Is it just me, or has peer review gotten more arduous lately? 4+ rounds of review isn't so unusual these days...

doi.org/10.1038/s415...
It was a long journey – 16 months from initial submission to acceptance. Is it just me, or has peer review gotten more arduous lately? 4+ rounds of review isn't so unusual these days...
Trust is lost not by scientists publishing evidence, but by dishonest actors misrepresenting the evidence.
www.theguardian.com/commentisfre...

Trust is lost not by scientists publishing evidence, but by dishonest actors misrepresenting the evidence.
www.theguardian.com/commentisfre...
The article itself contains multiple falsehoods and deep mischaracterizations.
Let's take every point in turn 👇🧵
www.nytimes.com/interactive/...
The article itself contains multiple falsehoods and deep mischaracterizations.
Let's take every point in turn 👇🧵
www.nytimes.com/interactive/...
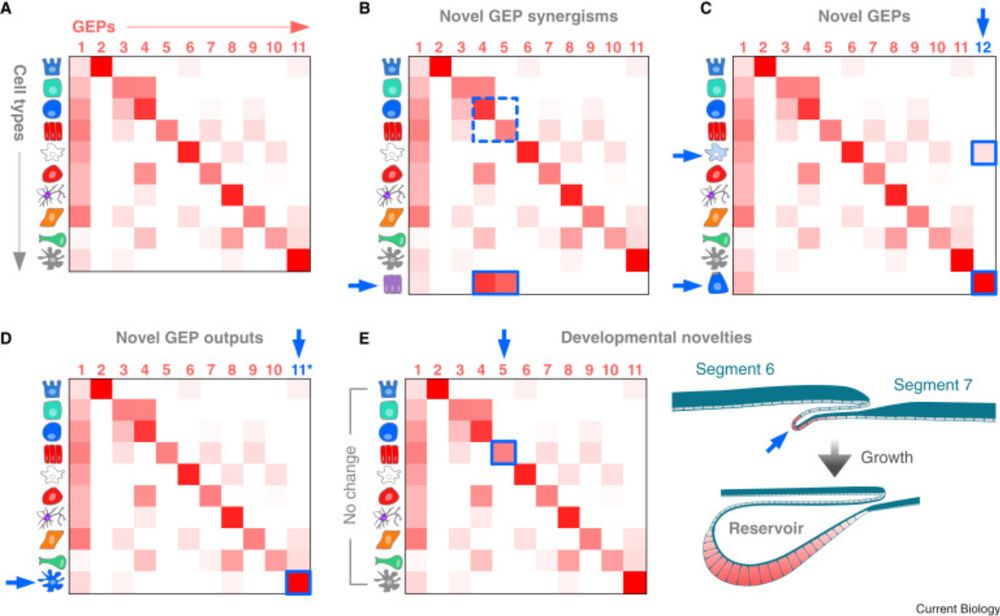
By Alison Barton, @mathiesoniain.bsky.social et al
www.nature.com/articles/s41...
By Alison Barton, @mathiesoniain.bsky.social et al
www.nature.com/articles/s41...
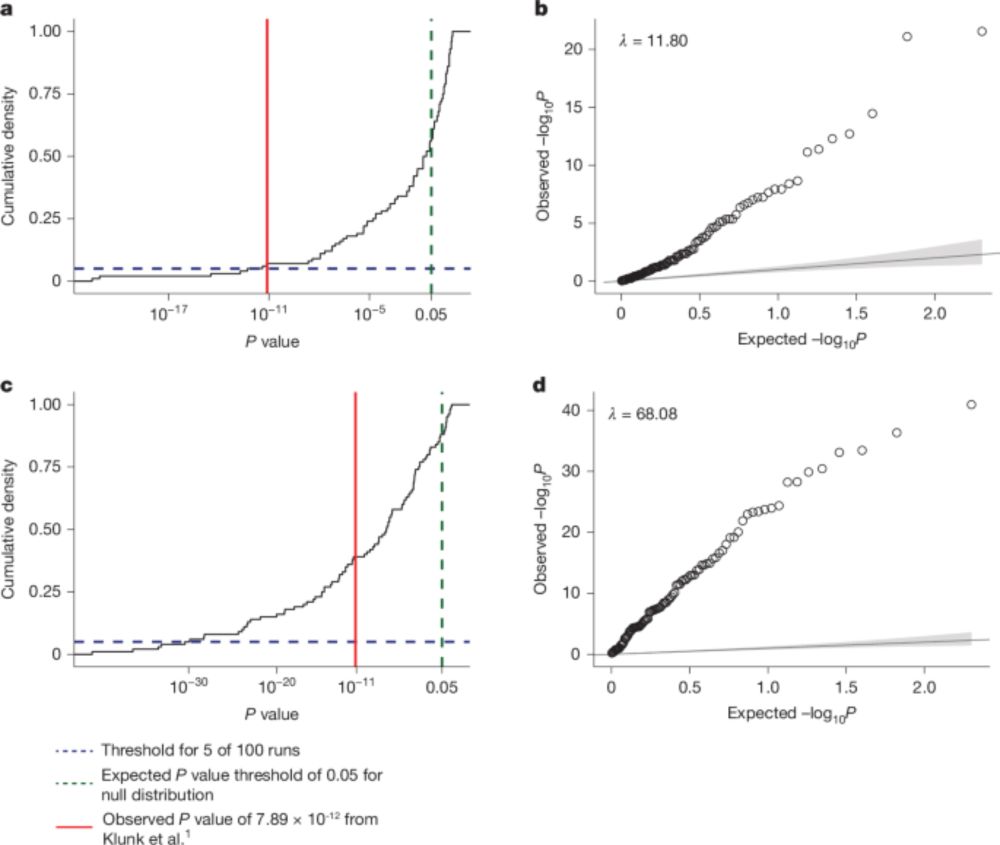
Led by Chaney Kalinich, Freddy Gonzalez, & Seth Redmond
www.biorxiv.org/content/10.1...

Led by Chaney Kalinich, Freddy Gonzalez, & Seth Redmond
www.biorxiv.org/content/10.1...
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...


Foto: Martin Surbeck, Kokolopori Research Project (thank you!).
🧪🧬 #popgen #evolution

Foto: Martin Surbeck, Kokolopori Research Project (thank you!).
🧪🧬 #popgen #evolution
www.cell.com/cell/fulltex...
“Zebrahub | Supp. Video 3: Light-sheet time-lapse of Tg(h2afva:h2afva-mcherry ; mezzo:eGFP) embryo starting at 50% epiboly.”
on #Vimeo vimeo.com/803719225
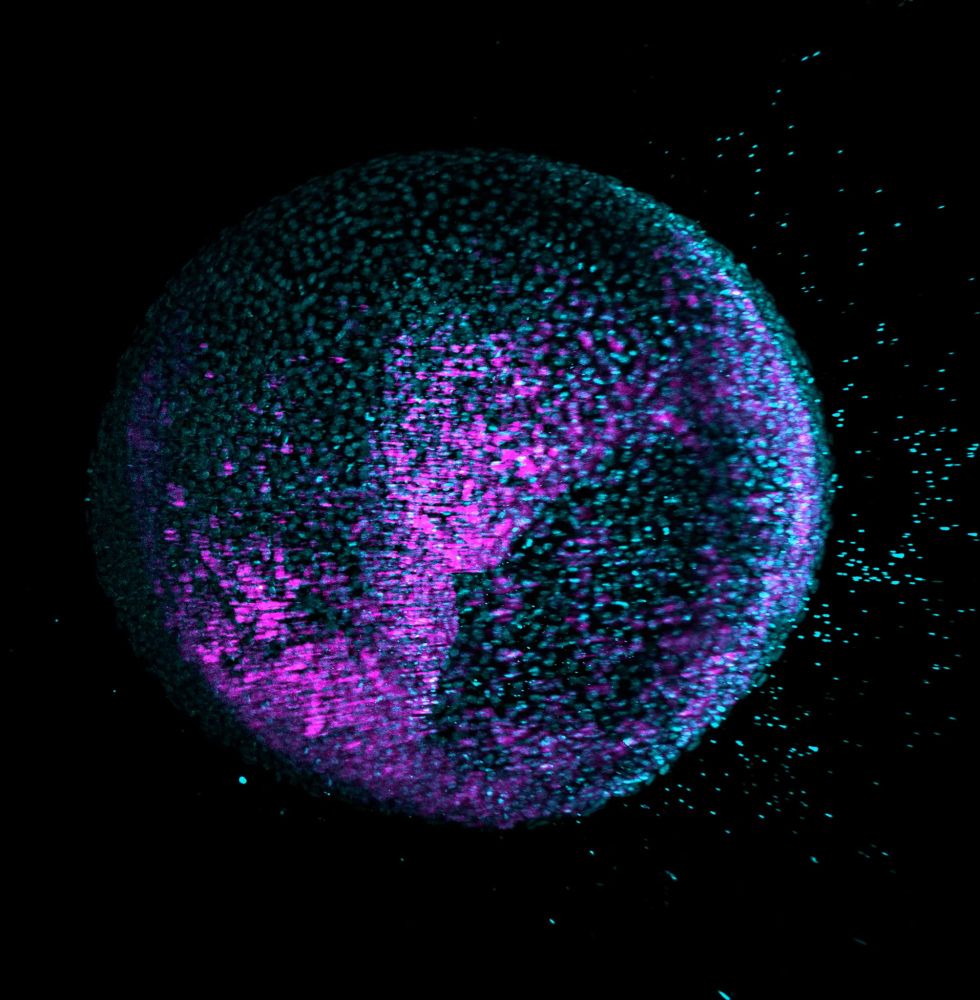
www.cell.com/cell/fulltex...
“Zebrahub | Supp. Video 3: Light-sheet time-lapse of Tg(h2afva:h2afva-mcherry ; mezzo:eGFP) embryo starting at 50% epiboly.”
on #Vimeo vimeo.com/803719225
They contain recognisable humanity, warmth and humour. Here's a thread of my favourites.


They contain recognisable humanity, warmth and humour. Here's a thread of my favourites.

1/7
openreview.net/forum?id=H7m...

1/7
openreview.net/forum?id=H7m...
Stay informed about the latest scverse events, software updates, and community news. Everything you need to know about foundational tools for single-cell omics analysis in one place.
go.bsky.app/UvFMa8d
Stay informed about the latest scverse events, software updates, and community news. Everything you need to know about foundational tools for single-cell omics analysis in one place.
go.bsky.app/UvFMa8d
How convinced are #SingleCell people by cell-cell communication methods? I'm still at "not very" 😅
I see them used everywhere, but I don't know of any really good way to validate them...
More importantly, I don't know of any *non-simulated* training data.
How convinced are #SingleCell people by cell-cell communication methods? I'm still at "not very" 😅
I see them used everywhere, but I don't know of any really good way to validate them...
More importantly, I don't know of any *non-simulated* training data.

~ squiggle
<- pretentious equals
; mouthless wink
%>% poop
! AHH (you have to shout it)
$ cha ching
[ subset
[[ subset and I really mean it!
(reposting an old tweet sparked by @amelia.mn)
~ squiggle
<- pretentious equals
; mouthless wink
%>% poop
! AHH (you have to shout it)
$ cha ching
[ subset
[[ subset and I really mean it!
(reposting an old tweet sparked by @amelia.mn)


