
@ucberkeleyofficial.bsky.social @innovativegenomics.bsky.social @hhmi.bsky.social
savagelab.org
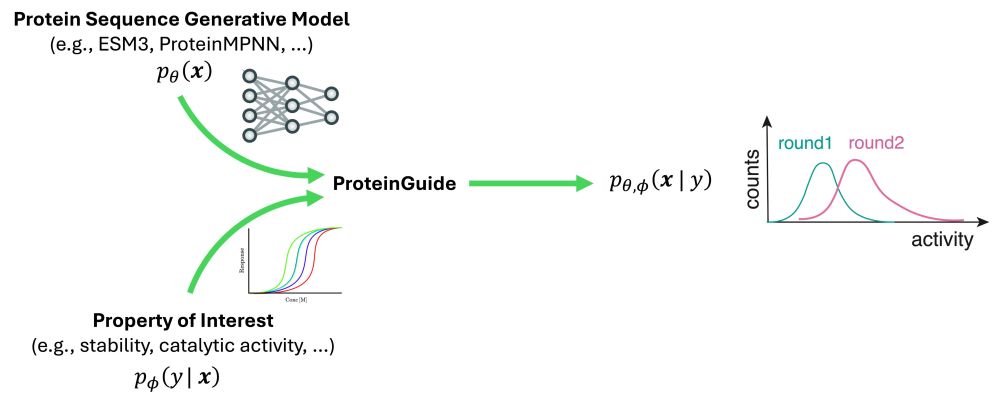


We noticed a pair of genes - a nuclease and a protease - shuffles between antiviral systems. We show how proteolysis activates the nuclease, triggering defense in known and unknown immune contexts.
tinyurl.com/2uwwy4ty


Muntathar Al-Shimary, @doudna-lab.bsky.social , @cresslab.bsky.social and I present a gene knockdown tool that works in diverse bacteriophages: CRISPRi through antisense RNA Targeting (CRISPRi-ART)!
Check it out @naturemicrobiol.bsky.social
www.nature.com/articles/s41...
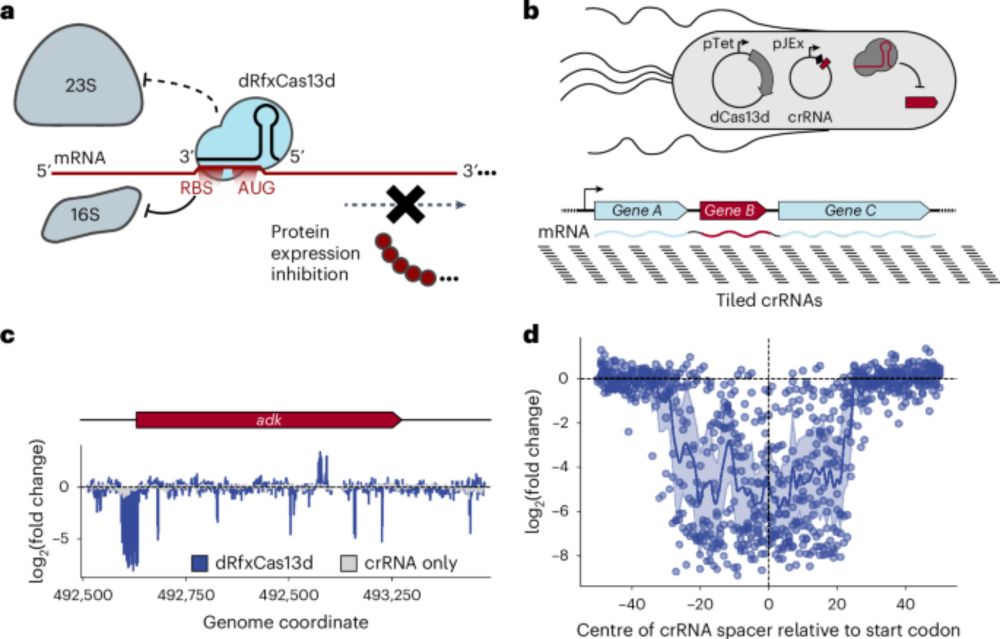
Muntathar Al-Shimary, @doudna-lab.bsky.social , @cresslab.bsky.social and I present a gene knockdown tool that works in diverse bacteriophages: CRISPRi through antisense RNA Targeting (CRISPRi-ART)!
Check it out @naturemicrobiol.bsky.social
www.nature.com/articles/s41...
Read more: ow.ly/t2JO50ULh10

Read more: ow.ly/t2JO50ULh10

Read more: ow.ly/t2JO50ULh10

Read more: ow.ly/t2JO50ULh10

