
Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...
@ycz23.bsky.social
arxiv.org/abs/2502.11333
(1/9)

@ycz23.bsky.social
www.nature.com/articles/s41...
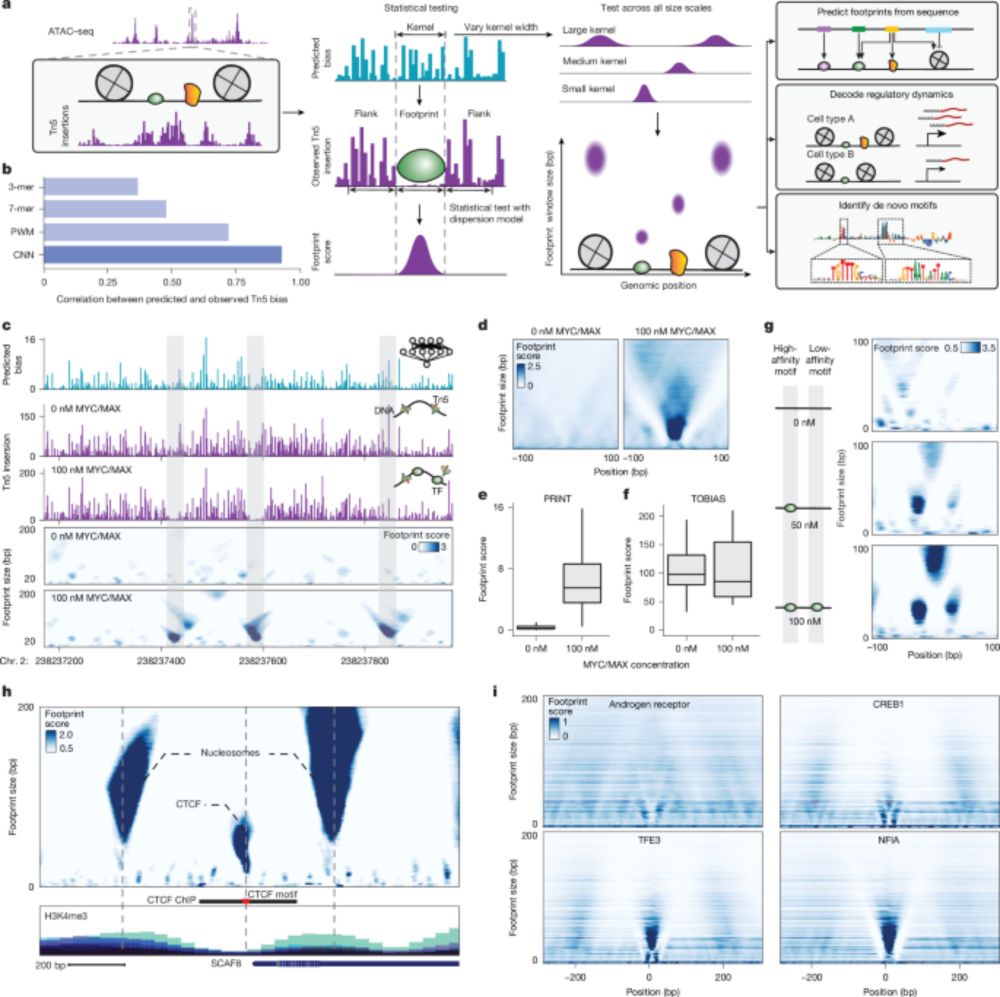
www.nature.com/articles/s41...
www.encodeproject.org/single-cell/...
www.encodeproject.org/single-cell/...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
(1/n) Ever wonder how transcription factors locate, "invade," and activate their enhancers during cell fate acquisition? 🧬🚀 Check out our latest collaborative work with the Soufi lab, now published in @Nature

(1/n) Ever wonder how transcription factors locate, "invade," and activate their enhancers during cell fate acquisition? 🧬🚀 Check out our latest collaborative work with the Soufi lab, now published in @Nature
Single-molecule states link transcription factor binding to gene expression
Deep insights into multiple facets of TF binding, accessibility & expression with biophysical models of single molecule data by Ben Doughty, Michaela Hinks, Bintu & Greenleaf labs

Single-molecule states link transcription factor binding to gene expression
Deep insights into multiple facets of TF binding, accessibility & expression with biophysical models of single molecule data by Ben Doughty, Michaela Hinks, Bintu & Greenleaf labs
www.nature.com/articles/s41...
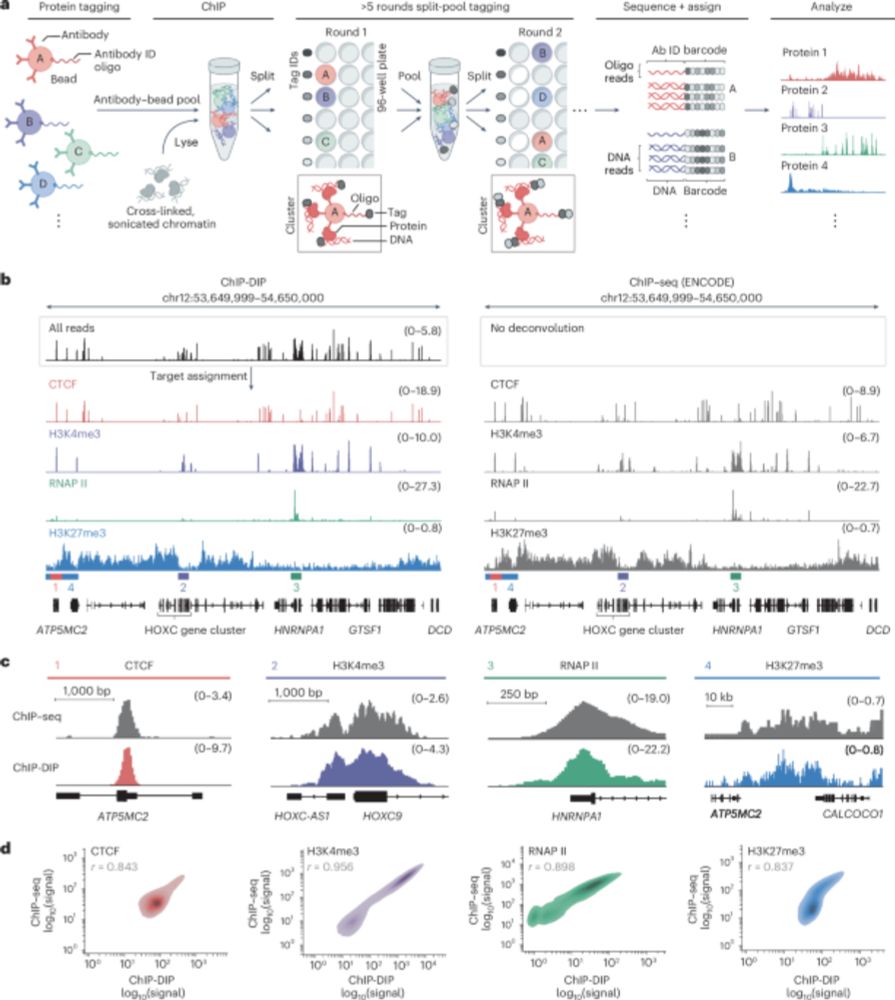
www.nature.com/articles/s41...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
If you are interested in doing a Ph.D. with me at UMass Chan Medical (Genomics and Comp Bio Department), see the links below. Deadline is Dec 1st.
If you are interested in doing a Ph.D. with me at UMass Chan Medical (Genomics and Comp Bio Department), see the links below. Deadline is Dec 1st.


