pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
#chemsky #compchem
doi.org/10.1021/acs....
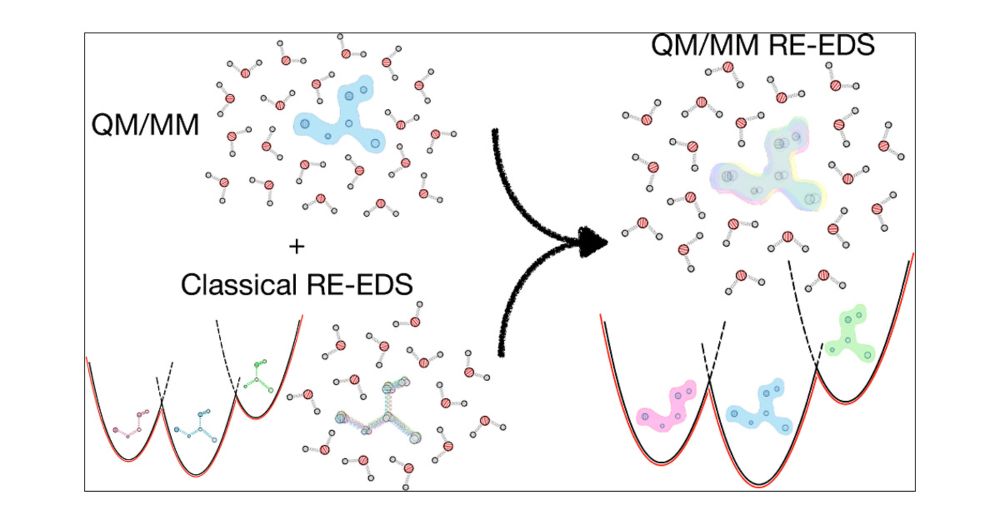
#chemsky #compchem
doi.org/10.1021/acs....
doi.org/10.1021/jacs...
#chemsky #compchem #opensource
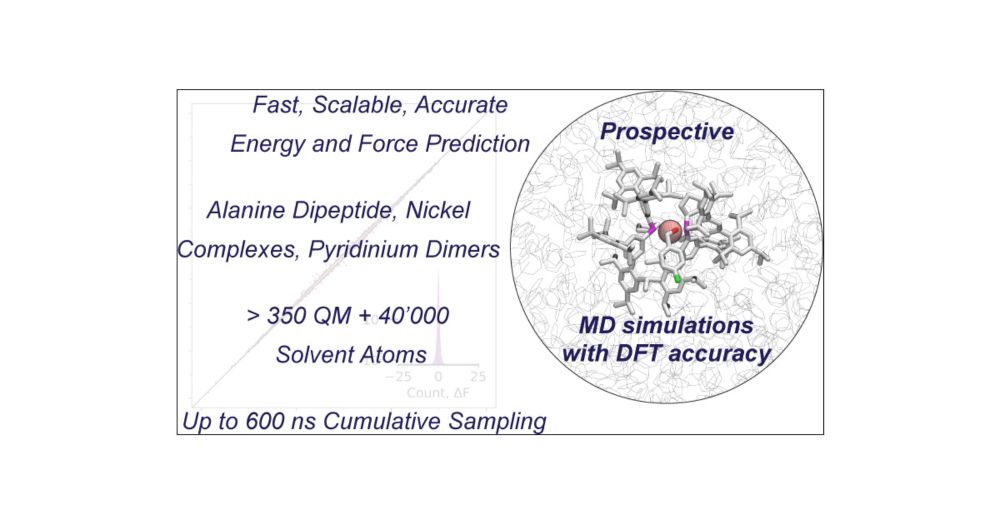
doi.org/10.1021/jacs...
#chemsky #compchem #opensource
chemrxiv.org/engage/chemr...

chemrxiv.org/engage/chemr...
pubs.rsc.org/en/content/a...
#chemsky
pubs.rsc.org/en/content/a...
#chemsky
#chemsky
pubs.acs.org/doi/10.1021/...
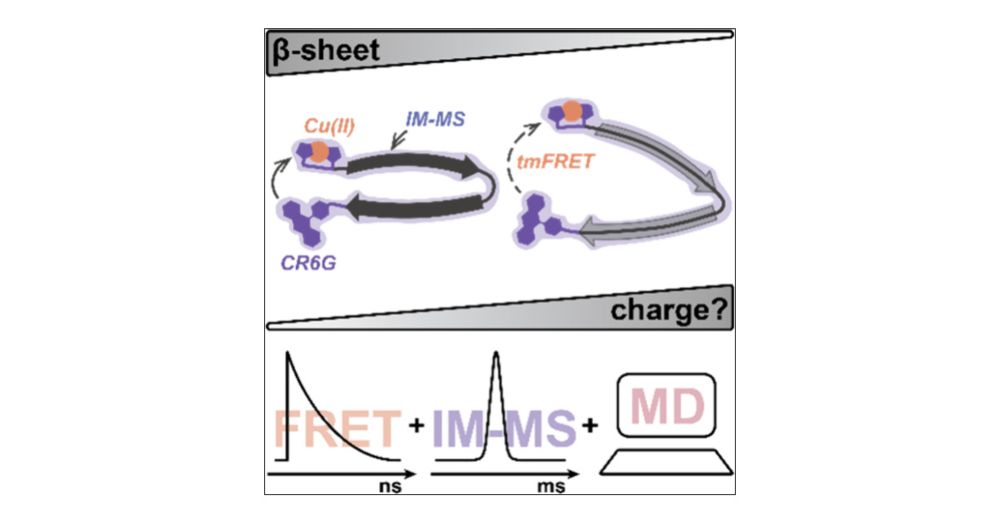
#chemsky
pubs.acs.org/doi/10.1021/...

doi.org/10.1063/5.01...
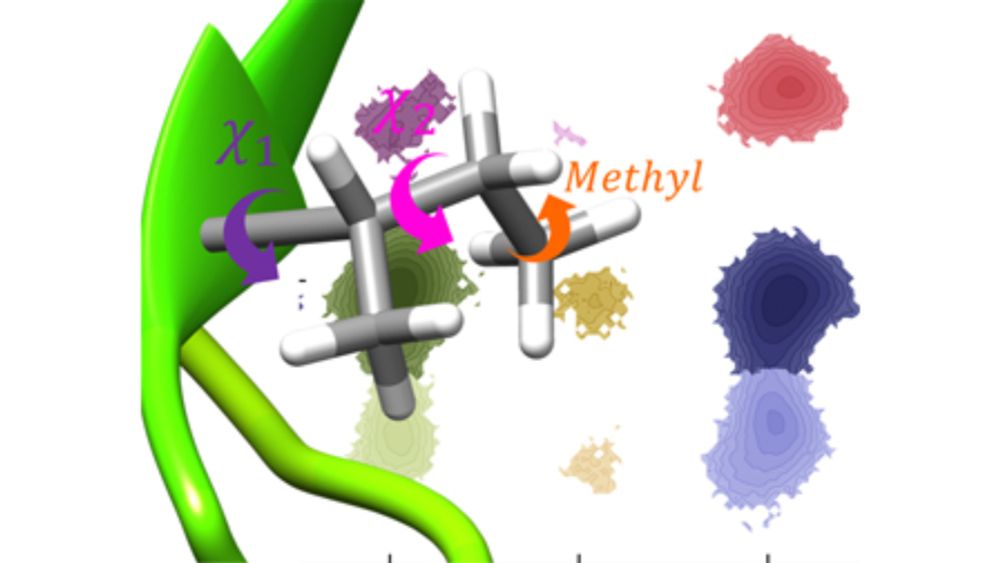
doi.org/10.1063/5.01...
pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...
doi.org/10.26434/che...

doi.org/10.26434/che...
link.springer.com/article/10.1...
#opensource code and data are in github.
github.com/rinikerlab/m...

link.springer.com/article/10.1...
#opensource code and data are in github.
github.com/rinikerlab/m...
chemrxiv.org/engage/chemr...

chemrxiv.org/engage/chemr...
As usual, it's #opensource, #opendata, and #openaccess
pubs.rsc.org/en/content/a...
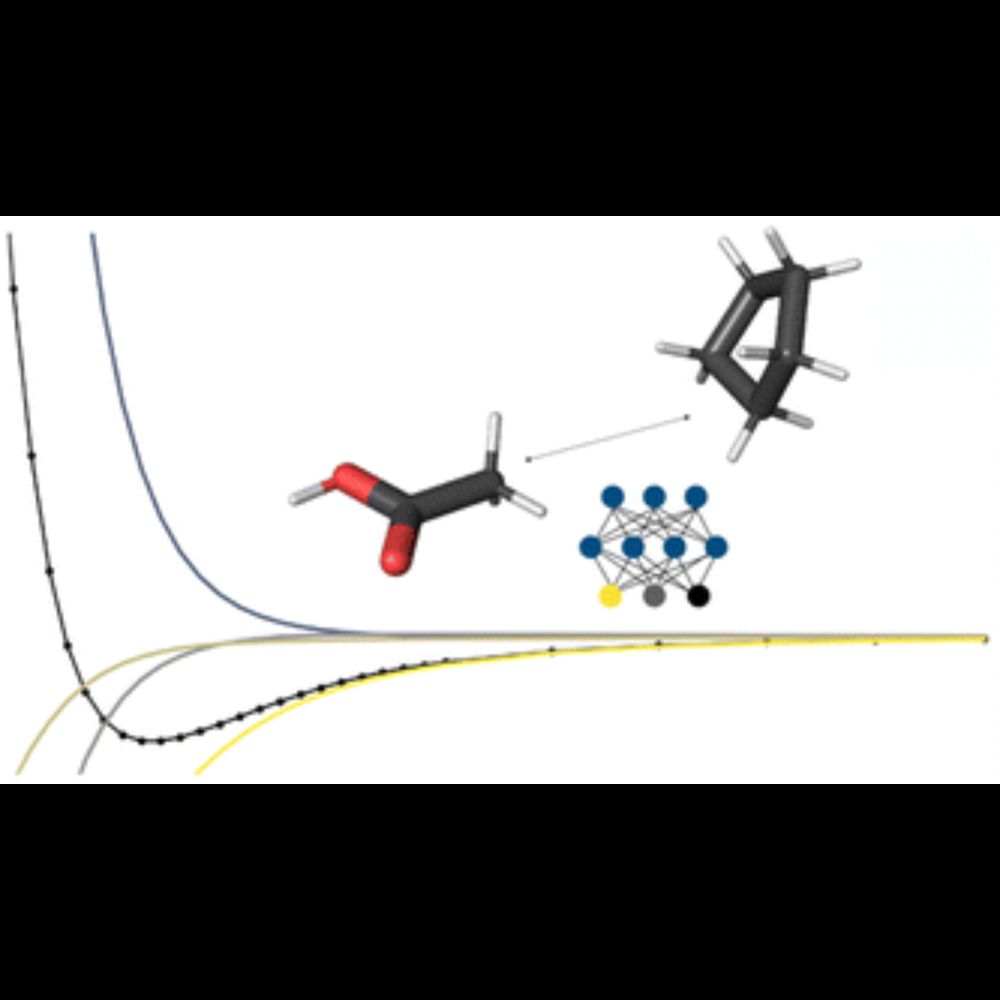
As usual, it's #opensource, #opendata, and #openaccess
pubs.rsc.org/en/content/a...
It's #opensource, #opendata, and #openaccess
pubs.acs.org/doi/10.1021/...
It's #opensource, #opendata, and #openaccess
pubs.acs.org/doi/10.1021/...

