🏳️🌈✡️ he/him
https://ryanzfriedman.com/
ryanzfriedman.com/2025/09/06/s...
ryanzfriedman.com/2025/09/06/s...
We answer the question: When you can synthesize any DNA sequence you want, how do you decide which ones are worth testing?
www.sciencedirect.com/science/arti...
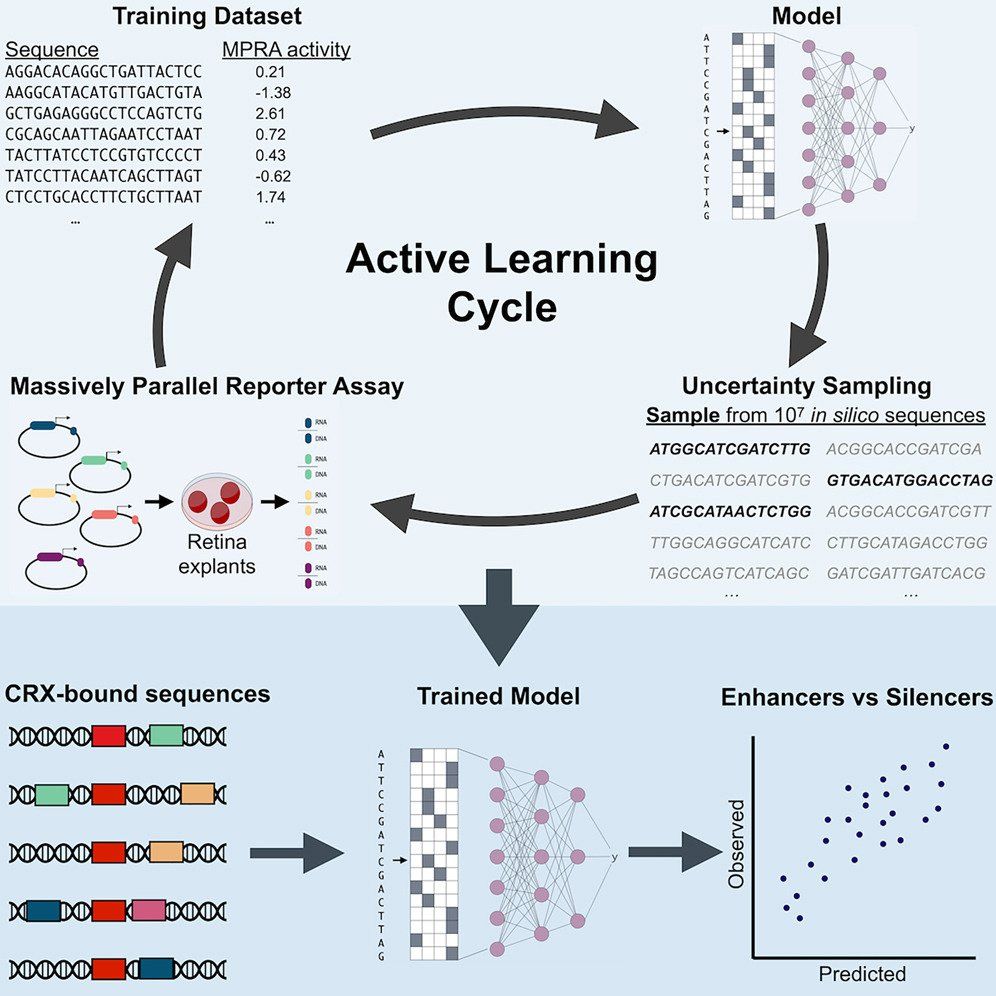
We answer the question: When you can synthesize any DNA sequence you want, how do you decide which ones are worth testing?
www.sciencedirect.com/science/arti...

The manuscript itself is also restructured. Figs 2 and 4 are swapped, there's a 5th fig for the K562 analysis, and we reworked the Discussion.
Apologies if threading isn't the way to go on Bsky. 🧬🔄
8/8
The manuscript itself is also restructured. Figs 2 and 4 are swapped, there's a 5th fig for the K562 analysis, and we reworked the Discussion.
Apologies if threading isn't the way to go on Bsky. 🧬🔄
8/8
These results show our model learns the context that distinguishes functionally non-equivalent motifs.
7/

These results show our model learns the context that distinguishes functionally non-equivalent motifs.
7/
Along with our other results, this shows active learning generates the data needed to learn regulatory grammars.
6/

Along with our other results, this shows active learning generates the data needed to learn regulatory grammars.
6/
5/

5/
4/
4/
This demonstrates that active learning is broadly effective and illustrate that enriching for active sequences is more informative
3/


This demonstrates that active learning is broadly effective and illustrate that enriching for active sequences is more informative
3/
2/


2/
TLDR: several new analyses, benchmarking w a 2nd MPRA dataset, and a refocused argument on active learning to leverage the capacity of MPRAs to generate large datasets.
www.biorxiv.org/content/10.1...
1/8
🧬🔄
TLDR: several new analyses, benchmarking w a 2nd MPRA dataset, and a refocused argument on active learning to leverage the capacity of MPRAs to generate large datasets.
www.biorxiv.org/content/10.1...
1/8
🧬🔄
journals.plos.org/ploscompbiol...
journals.plos.org/ploscompbiol...
Is there anyone in genomics 🖥️🧬 who wants to share how they organize their reference managers? I'm looking for more meaningful and detailed categories.
Is there anyone in genomics 🖥️🧬 who wants to share how they organize their reference managers? I'm looking for more meaningful and detailed categories.


