In the largest study of its kind (402k UKB individuals; 7 continuous traits + 3 diseases), we asked: If your phenotype deviates from common-variant polygenic score prediction, what's driving that difference?
www.medrxiv.org/content/10.6...

In the largest study of its kind (402k UKB individuals; 7 continuous traits + 3 diseases), we asked: If your phenotype deviates from common-variant polygenic score prediction, what's driving that difference?
www.medrxiv.org/content/10.6...

We combined large-scale human genetics with CRISPR-Cas9 editing in fat cells to identify genes linked to fat accumulation.
Check out the full study, now published in AJHG! www.cell.com/ajhg/fulltex...

We combined large-scale human genetics with CRISPR-Cas9 editing in fat cells to identify genes linked to fat accumulation.
Check out the full study, now published in AJHG! www.cell.com/ajhg/fulltex...
More info here: bit.ly/biomedDPhil
🗓️ App deadline: June 30th 2025.
Please share!
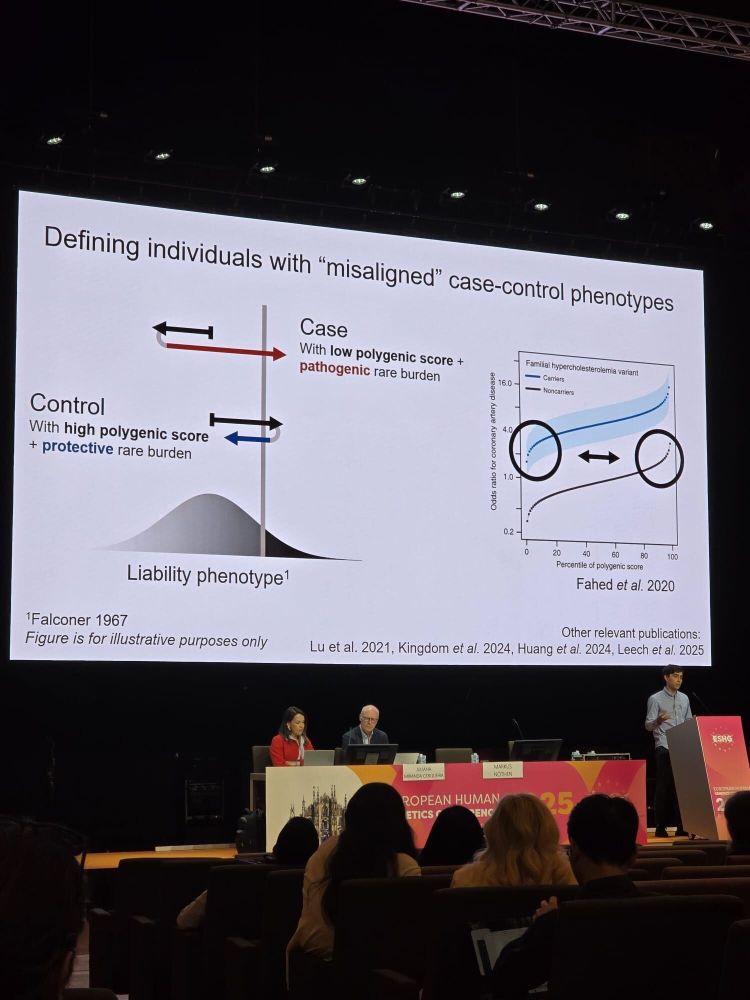
I presented our work demonstrating that individuals whose phenotype deviates from genetic expectation are enriched for rare damaging variants.
This has implications for:
- Screening of rare disorders 🔎
- Target discovery 💊
- Improving trait prediction 📈

I presented our work demonstrating that individuals whose phenotype deviates from genetic expectation are enriched for rare damaging variants.
This has implications for:
- Screening of rare disorders 🔎
- Target discovery 💊
- Improving trait prediction 📈

