Nathan Salomonis
@nathansalomonis.bsky.social
Computational biology and splicing regulation
New in STM, check out our integrative bulk & single-cell long-read analysis to discover and test new therapies in leukemia. We developed OncoSplice, to define new splicing defined subtypes and predict regulators. PRMT5 inhibition partially rescued mis-splicing & inhibited leukemic growth. More soon!
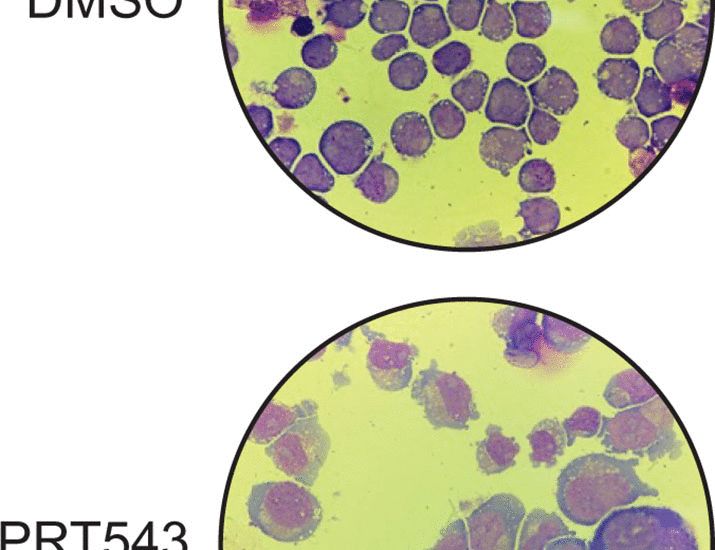
Splicing regulatory dynamics for precision analysis and treatment of heterogeneous leukemias
Broad dysregulated splicing in AML partially phenocopies splicing factor mutations, is prognostic, and can be pharmacologically reversed.
www.science.org
May 7, 2025 at 11:05 PM
New in STM, check out our integrative bulk & single-cell long-read analysis to discover and test new therapies in leukemia. We developed OncoSplice, to define new splicing defined subtypes and predict regulators. PRMT5 inhibition partially rescued mis-splicing & inhibited leukemic growth. More soon!
Reposted by Nathan Salomonis
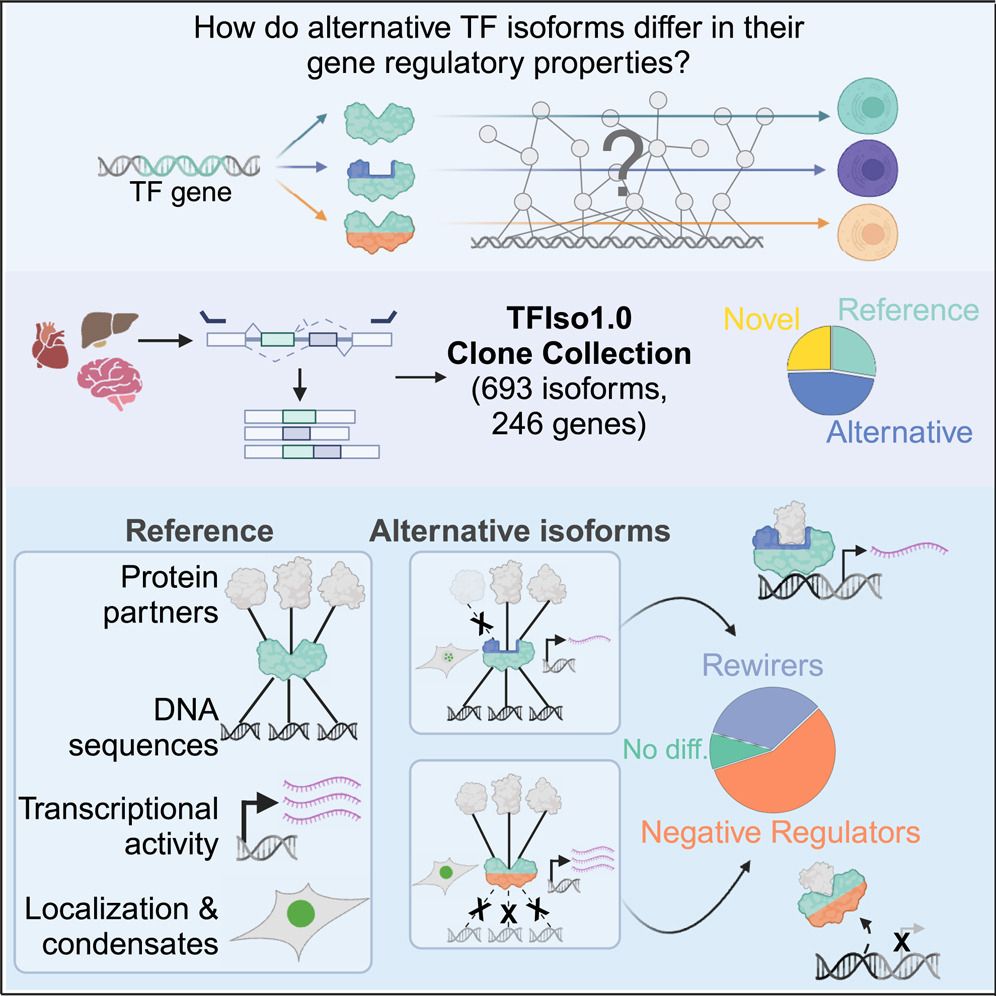
our work on the molecular differences between transcription factor isoforms is out now in Molecular Cell!
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
March 26, 2025 at 5:11 PM
our work on the molecular differences between transcription factor isoforms is out now in Molecular Cell!
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
Reposted by Nathan Salomonis
New algorithm for spatial transcriptome analysis that predicts the crosstalk of cell types co-localized in tissue niches. Also studies the downstream effects of cell-cell interactions by inferring covarying gene programs.
doi.org/10.1038/s414...
#spatial #transcriptomics #scrnaseq #NMF
doi.org/10.1038/s414...
#spatial #transcriptomics #scrnaseq #NMF
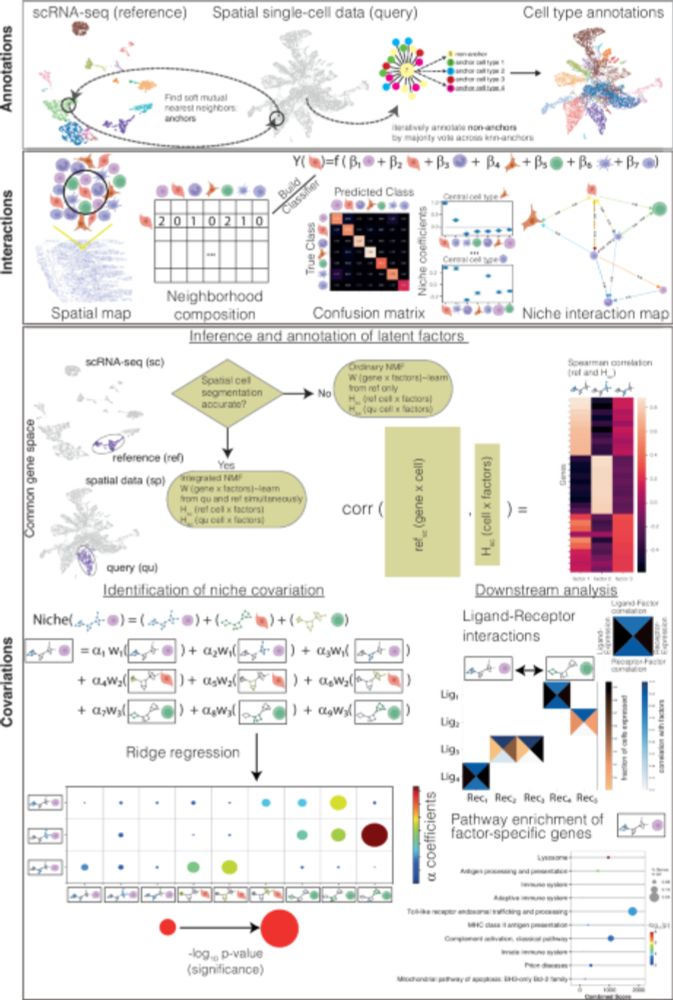
NiCo identifies extrinsic drivers of cell state modulation by niche covariation analysis - Nature Communications
A key question in single-cell biology is how cells communicate and exchange information with neighboring cells in tissues. Here, the authors introduce NiCo to predict the downstream effect of cell-cel...
doi.org
December 8, 2024 at 4:09 AM
New algorithm for spatial transcriptome analysis that predicts the crosstalk of cell types co-localized in tissue niches. Also studies the downstream effects of cell-cell interactions by inferring covarying gene programs.
doi.org/10.1038/s414...
#spatial #transcriptomics #scrnaseq #NMF
doi.org/10.1038/s414...
#spatial #transcriptomics #scrnaseq #NMF
Reposted by Nathan Salomonis
Major milestone: Tabula Sapiens 2.0 maps tissue composition & TF expression in 175 cell types, identifying 745 ubiquitous & 890 cell type-specific TFs (many still uncharacterized -> #Codebook) w/ roles in tissue homeostasis, stress response & metabolism. #SingleCell www.biorxiv.org/content/10.1...

Tabula Sapiens reveals transcription factor expression, senescence effects, and sex-specific features in cell types from 28 human organs and tissues
The Tabula Sapiens is a reference human cell atlas containing single cell transcriptomic data from more than two dozen organs and tissues. Here we report Tabula Sapiens 2.0 which includes data from ni...
www.biorxiv.org
December 8, 2024 at 8:55 AM
Major milestone: Tabula Sapiens 2.0 maps tissue composition & TF expression in 175 cell types, identifying 745 ubiquitous & 890 cell type-specific TFs (many still uncharacterized -> #Codebook) w/ roles in tissue homeostasis, stress response & metabolism. #SingleCell www.biorxiv.org/content/10.1...
Reposted by Nathan Salomonis
Super excited to share our Human Neural Organoid Atlas, now out in Nature! Led by @zhisonghe.bsky.social @josch1.bsky.social, and myself, this resource was created from 36 scRNA-seq datasets—totalling over 1.7 million cells! 🔬✨
www.nature.com/articles/s41...
Find out how it can serve you ⏬
🧵1/8
Find out how it can serve you ⏬
🧵1/8
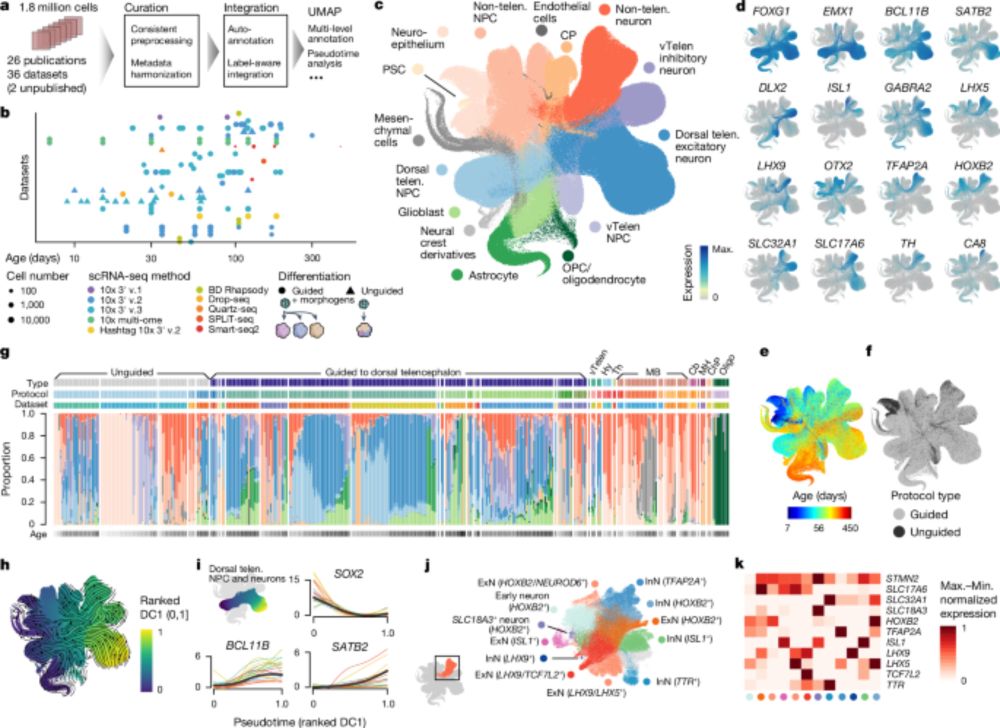
An integrated transcriptomic cell atlas of human neural organoids - Nature
A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, sho...
www.nature.com
November 21, 2024 at 10:11 AM
Super excited to share our Human Neural Organoid Atlas, now out in Nature! Led by @zhisonghe.bsky.social @josch1.bsky.social, and myself, this resource was created from 36 scRNA-seq datasets—totalling over 1.7 million cells! 🔬✨
www.nature.com/articles/s41...
Find out how it can serve you ⏬
🧵1/8
Find out how it can serve you ⏬
🧵1/8

