
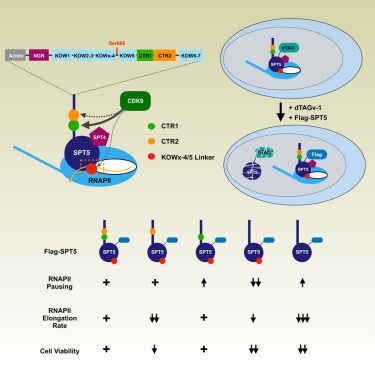
New chromatin state, transcription resistive, different from activation & silencing, marked by H3K36me, needs interplay between SDG8 and other H3K36me MTs.
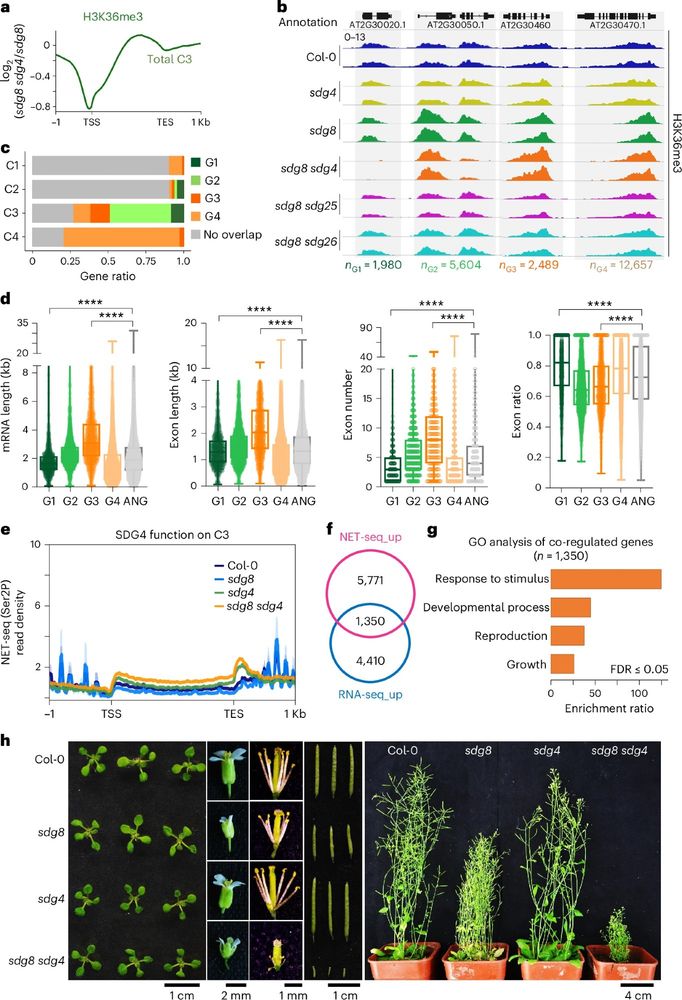
New chromatin state, transcription resistive, different from activation & silencing, marked by H3K36me, needs interplay between SDG8 and other H3K36me MTs.


Join Peter Kindgren's group @umeaplantscicentre.bsky.social and generate superior plants in terms of biomass and stress resilience.
Read more here and apply:
slu.se/en/about-slu...
#PlantSciJobs #PlantSciJob #hiring #plantscience
Join Peter Kindgren's group @umeaplantscicentre.bsky.social and generate superior plants in terms of biomass and stress resilience.
Read more here and apply:
slu.se/en/about-slu...
#PlantSciJobs #PlantSciJob #hiring #plantscience

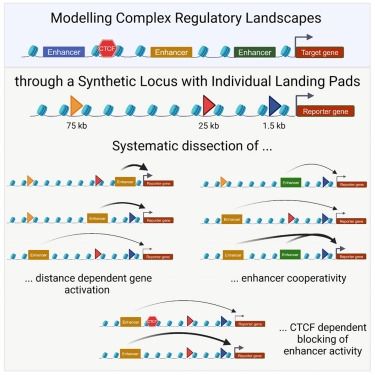

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
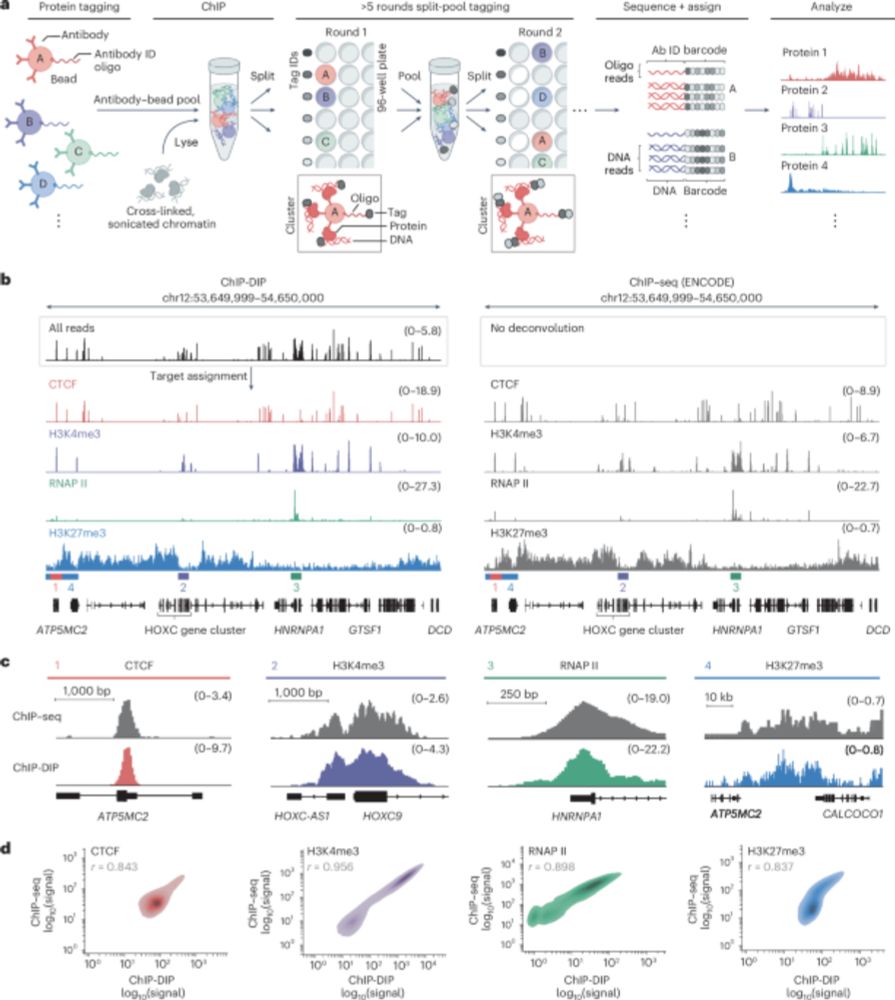
www.nature.com/articles/s41...

prideinstem.org/lgbtstemday/

prideinstem.org/lgbtstemday/
What is your favorite RNase inhibitor for plant RNAses? More precisely it's for young tomato 🍅 fruits.
#RNA #RNase
Thank you 🙏🙏
What is your favorite RNase inhibitor for plant RNAses? More precisely it's for young tomato 🍅 fruits.
#RNA #RNase
Thank you 🙏🙏


