We develop cutting-edge technologies to explore genome evolution!
Lab Website: https://schneebergerlab.org/
A platform-agnostic framework for single-cell recombination analysis. We study crossover variation in mutants & natural lines, identifying the largest natural inversion in A. thaliana to date!
📄 doi.org/10.64898/202...
🐱 github.com/schneeberger...
A platform-agnostic framework for single-cell recombination analysis. We study crossover variation in mutants & natural lines, identifying the largest natural inversion in A. thaliana to date!
📄 doi.org/10.64898/202...
🐱 github.com/schneeberger...
🌱 An extreme meiotic outcome: faithful segregation without crossovers in a holocentric plant.
🔗 doi.org/10.64898/202...
#Preprint #GenomeEvolution #Meiosis4eva #centromere #Pangenomics
🌱 An extreme meiotic outcome: faithful segregation without crossovers in a holocentric plant.
🔗 doi.org/10.64898/202...
#Preprint #GenomeEvolution #Meiosis4eva #centromere #Pangenomics
Together, they connect centromere repeat evolution, karyotype dynamics, and meiotic recombination outcomes, revealing how holocentric genomes evolve and function. 🧬👇

Together, they connect centromere repeat evolution, karyotype dynamics, and meiotic recombination outcomes, revealing how holocentric genomes evolve and function. 🧬👇
Love genome plasticity, computational methods, and solving big questions in plant biology? Join our newly established Institute for Crop Biology at HHU Düsseldorf.
More info on schneebergerlab.org/career/
Apply by Nov. 30 | 3-year position
Love genome plasticity, computational methods, and solving big questions in plant biology? Join our newly established Institute for Crop Biology at HHU Düsseldorf.
More info on schneebergerlab.org/career/
Apply by Nov. 30 | 3-year position
Complexity welcome: Pangenome graphs for comprehensive population genomics
#pangenomes #plantscience #genomegraphs
www.cambridge.org/core/journal...

Complexity welcome: Pangenome graphs for comprehensive population genomics
#pangenomes #plantscience #genomegraphs
www.cambridge.org/core/journal...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Check out this thread from Xiao Dong to learn more.
🚨 My first PhD preprint is out on bioRxiv!
We show how tiny mutations—point mutations and kb-level indels—can shape the massive structure of Arabidopsis centromeres.
📄 Read it here: doi.org/10.1101/2025...
🧵 A short thread 👇
Check out this thread from Xiao Dong to learn more.
➡️ bit.ly/42Mys6F

➡️ bit.ly/42Mys6F
Check out our blog post and the latest @naturepodcast.bsky.social episode!
📝 go.nature.com/3G73vT7
🎙️ www.nature.com/articles/d41...

Check out our blog post and the latest @naturepodcast.bsky.social episode!
📝 go.nature.com/3G73vT7
🎙️ www.nature.com/articles/d41...
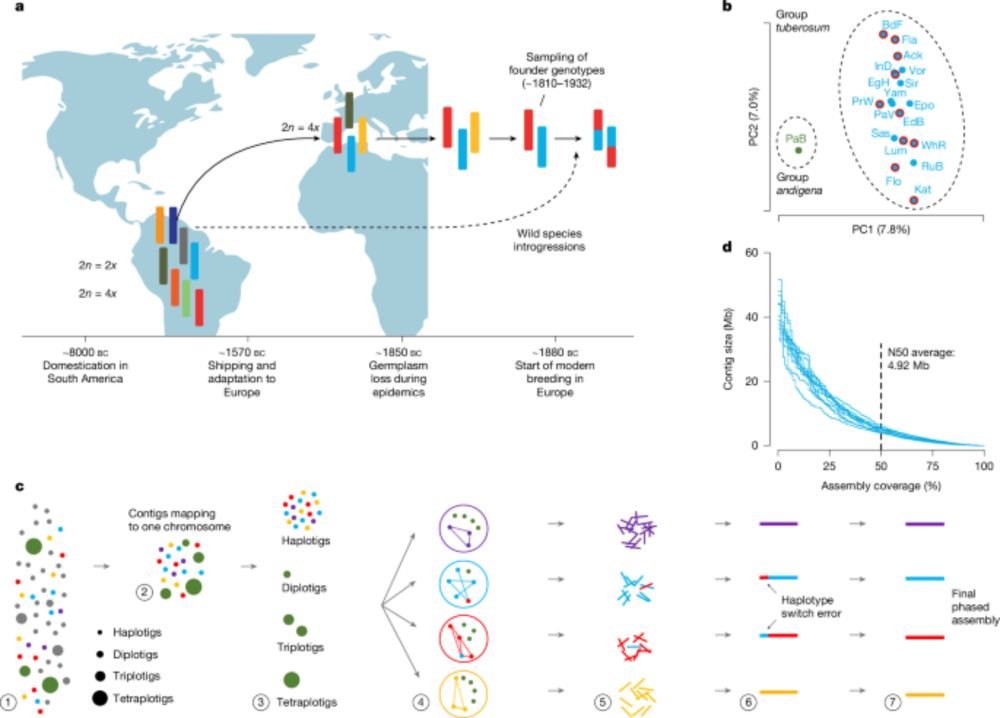
We present the phased pan-genome of tetraploid European potato, based on 10 historical cultivars representing 85% of European potato diversity.
Learn more below!
🔗 www.nature.com/articles/s41...
1/7
We present the phased pan-genome of tetraploid European potato, based on 10 historical cultivars representing 85% of European potato diversity.
Learn more below!
🔗 www.nature.com/articles/s41...
1/7

