In this preprint, we compare the differences when Mycobacterial samples are sequenced with either long- or short-read sequencing and show that long-read sequencing (as exemplified by Oxford Nanopore Technologies) is now…
In this preprint, we compare the differences when Mycobacterial samples are sequenced with either long- or short-read sequencing and show that long-read sequencing (as exemplified by Oxford Nanopore Technologies) is now…
Zam Iqbal, at the University of Bath, led this epic study published today in Nature Methods where we assembled all SARS-CoV-2 genomes deposited in the European Nucleotide Archive before 2 March 2023. In total a…

Zam Iqbal, at the University of Bath, led this epic study published today in Nature Methods where we assembled all SARS-CoV-2 genomes deposited in the European Nucleotide Archive before 2 March 2023. In total a…
Pleased and excited to be a small part of the Gram-Negative Antibiotic Discovery Innovator (Gr-ADI) consortium which was announced today. Gr-ADI is a join initiative between the Gates Foundation, Novo Nordisk Foundation, and Wellcome. Our project is headed by Dr Annette…
Pleased and excited to be a small part of the Gram-Negative Antibiotic Discovery Innovator (Gr-ADI) consortium which was announced today. Gr-ADI is a join initiative between the Gates Foundation, Novo Nordisk Foundation, and Wellcome. Our project is headed by Dr Annette…
Viki successfully defended her DPhil thesis on Tuesday 9 December 2025 - well done! I always tell everyone they will enjoy the viva after half an hour or so and I hope she did. Her examiners were Professor Tim Walker and Associate Professor Conor Meehan.

Viki successfully defended her DPhil thesis on Tuesday 9 December 2025 - well done! I always tell everyone they will enjoy the viva after half an hour or so and I hope she did. Her examiners were Professor Tim Walker and Associate Professor Conor Meehan.
This paper just published in Microbial Genomics examines how well our software tool, gnomonicus, predicts to which antibiotics a clinical sample that has been whole-genome sequenced is resistant. To do so, it implements the second…

This paper just published in Microbial Genomics examines how well our software tool, gnomonicus, predicts to which antibiotics a clinical sample that has been whole-genome sequenced is resistant. To do so, it implements the second…
Research into how resistance emerges, spreads and can be prevented is vital. Our group works to understand these processes and develop better tools to detect, monitor and combat antimicrobial resistance.
#WAAW @oxfordbrc.bsky.social

Research into how resistance emerges, spreads and can be prevented is vital. Our group works to understand these processes and develop better tools to detect, monitor and combat antimicrobial resistance.
#WAAW @oxfordbrc.bsky.social
In previous work we've used "traditional" machine-learning approaches, like XGBoost, to learn and therefore predict which mutations in PncA confer resistance to pyrazinamide, one of the four…

In previous work we've used "traditional" machine-learning approaches, like XGBoost, to learn and therefore predict which mutations in PncA confer resistance to pyrazinamide, one of the four…
Ruan Spies did a careful systematic analysis of the publicly-available pipelines that claimed to process raw genetics files from M. tuberculosis complex samples, including the pipeline available via EIT's GPAS which we have written. The…
Ruan Spies did a careful systematic analysis of the publicly-available pipelines that claimed to process raw genetics files from M. tuberculosis complex samples, including the pipeline available via EIT's GPAS which we have written. The…
Received a lovely email from Dr Peter Cotgreave who is the Chief Executive of the Microbiological Society to say Dylan's manuscript where he created a resistance catalogue for bedaquiline was one of the most viewed…
Received a lovely email from Dr Peter Cotgreave who is the Chief Executive of the Microbiological Society to say Dylan's manuscript where he created a resistance catalogue for bedaquiline was one of the most viewed…
bit.ly/47rwf42
@modmedmicro.bsky.social
@oxfordbrc.bsky.social

bit.ly/47rwf42
@modmedmicro.bsky.social
@oxfordbrc.bsky.social
Last Thursday this work which we'd previously preprinted looking at looking at rifampicin-resistant subpopulations in clinical M. tuberculosis samples was published in JAC-Antimicrobial Resistance. If you want to know more…
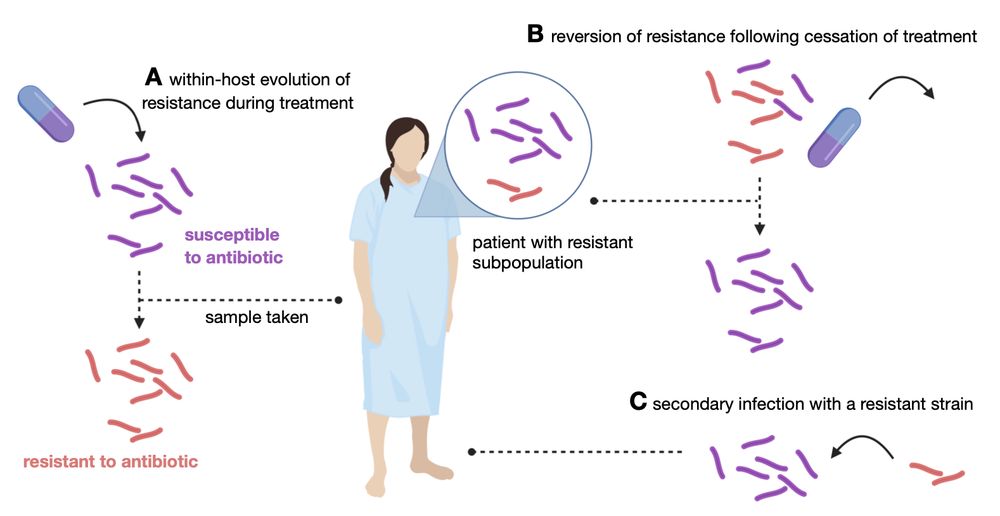
Last Thursday this work which we'd previously preprinted looking at looking at rifampicin-resistant subpopulations in clinical M. tuberculosis samples was published in JAC-Antimicrobial Resistance. If you want to know more…
The CRyPTIC project carried out many exciting research projects but it never quite got around to building a catalogue of resistance- and susceptible-associated genetic variants in M. tuberculosis, in part…

The CRyPTIC project carried out many exciting research projects but it never quite got around to building a catalogue of resistance- and susceptible-associated genetic variants in M. tuberculosis, in part…
This work was carried out by Xibei Zhang, who is doing her PhD with Peter Coveney at UCL. It builds on earlier work I did using alchemical free energy methods to calculate whether individual mutations in the protein target of…

This work was carried out by Xibei Zhang, who is doing her PhD with Peter Coveney at UCL. It builds on earlier work I did using alchemical free energy methods to calculate whether individual mutations in the protein target of…
The CRyPTIC project ran from 2016 to 2022 and collected >20,000 clinical samples from patients with tuberculosis. Each sample underwent whole genome sequencing and also was inoculated onto a 96-well plate containing 13 different antibiotics at a range…

The CRyPTIC project ran from 2016 to 2022 and collected >20,000 clinical samples from patients with tuberculosis. Each sample underwent whole genome sequencing and also was inoculated onto a 96-well plate containing 13 different antibiotics at a range…
Most of us attended the third Diagnostics for Low- and Middle-Income Countries (Dx4LMICs) conference at Reuben College in Oxford over the last two days. I am a Fellow at Reuben College and helped our President, Professor Lord Lionel Tarassenko and several others,…

Most of us attended the third Diagnostics for Low- and Middle-Income Countries (Dx4LMICs) conference at Reuben College in Oxford over the last two days. I am a Fellow at Reuben College and helped our President, Professor Lord Lionel Tarassenko and several others,…
The CRyPTIC project ran from 2016 to 2022 and collected >20,000 clinical samples from patients with tuberculosis. Each sample underwent whole genome sequencing and also was inoculated onto a 96-well plate containing 13 different antibiotics at a range…

The CRyPTIC project ran from 2016 to 2022 and collected >20,000 clinical samples from patients with tuberculosis. Each sample underwent whole genome sequencing and also was inoculated onto a 96-well plate containing 13 different antibiotics at a range…
Thanks to funding from the OxCoD4TB project, I am hiring a postdoctoral research associate. This consortium pulls together expertise from all over the University, including in diagnostics, data science, drug and vaccine design, preclinical testing and clinical testing and brings it to…
Thanks to funding from the OxCoD4TB project, I am hiring a postdoctoral research associate. This consortium pulls together expertise from all over the University, including in diagnostics, data science, drug and vaccine design, preclinical testing and clinical testing and brings it to…
Our paper describing how a convolutional neural network model can determine the minimum inhibitory concentrations (MICs) from a photograph of the 96-well plate after two weeks incubation has been published in the…
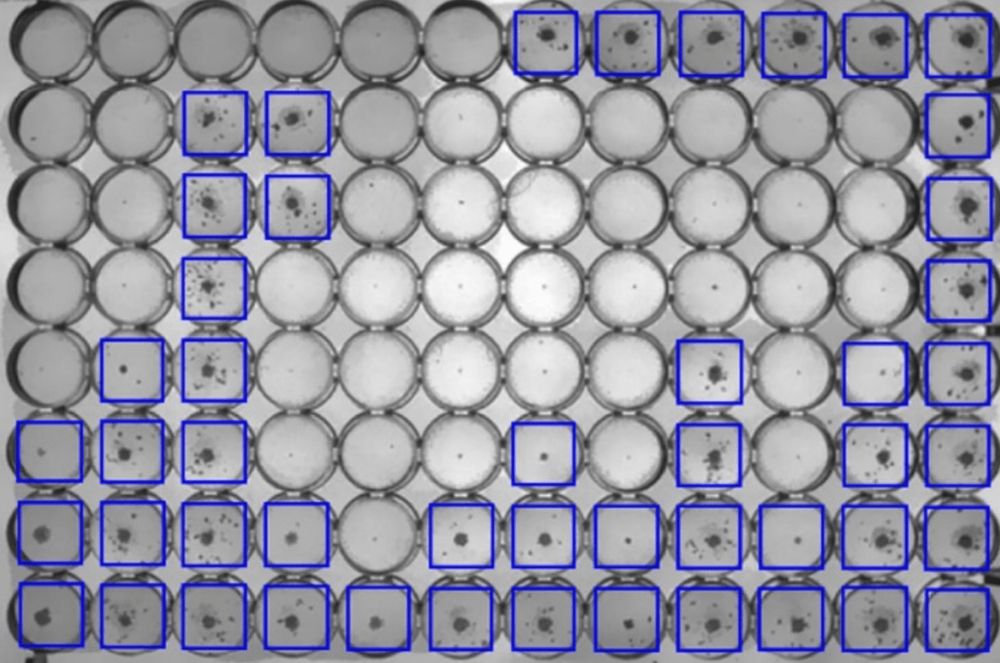
Our paper describing how a convolutional neural network model can determine the minimum inhibitory concentrations (MICs) from a photograph of the 96-well plate after two weeks incubation has been published in the…
Senior Biostatistical Researcher bit.ly/4kRfZxI
Senior Bioinformatic Engineer bit.ly/4ecl0hF
both Grade 8 or 7, closing noon Wed 2 Jul
Computational Mycobacteriologist
Grade 7, closing noon Fri 4 Jul bit.ly/4ndKOhF
@medsci.ox.ac.uk
Senior Biostatistical Researcher bit.ly/4kRfZxI
Senior Bioinformatic Engineer bit.ly/4ecl0hF
both Grade 8 or 7, closing noon Wed 2 Jul
Computational Mycobacteriologist
Grade 7, closing noon Fri 4 Jul bit.ly/4ndKOhF
@medsci.ox.ac.uk
Several of us attended the 45th Annual Congress of the European Society of Mycobacteriology in Lisbon, Portugal. This is probably my favourite scientific meeting -- small, friendly and with plenty of time to talk, including organised social events. Viki Brunner, Dylan…

Several of us attended the 45th Annual Congress of the European Society of Mycobacteriology in Lisbon, Portugal. This is probably my favourite scientific meeting -- small, friendly and with plenty of time to talk, including organised social events. Viki Brunner, Dylan…
53,897 clinical tuberculosis samples with genetics and drug susceptibility testing data!

53,897 clinical tuberculosis samples with genetics and drug susceptibility testing data!
Come and help @philipwfowler.bsky.social use the large TB datasets we hold to help the OxCoD4TB consortium develop new therapeutics for tuberculosis.
Grade 7, funding until 28 Feb 2028, deadline noon Fri 4 July
bit.ly/4ndKOhF
Come and help @philipwfowler.bsky.social use the large TB datasets we hold to help the OxCoD4TB consortium develop new therapeutics for tuberculosis.
Grade 7, funding until 28 Feb 2028, deadline noon Fri 4 July
bit.ly/4ndKOhF
0906 @vbrunner.bsky.social “RIF and subpopulations in TB”
0918 Ruan Spies “comparing TB pipelines" both in Hall 5
1000 Nicole Stoesser “plasmids & 1Health” in H2
1509 Jane Wei in Hall 2 “CAP and EHR”
P1405 - Valentina Pennetta
P2242 - Junko Takada
0906 @vbrunner.bsky.social “RIF and subpopulations in TB”
0918 Ruan Spies “comparing TB pipelines" both in Hall 5
1000 Nicole Stoesser “plasmids & 1Health” in H2
1509 Jane Wei in Hall 2 “CAP and EHR”
P1405 - Valentina Pennetta
P2242 - Junko Takada

