studying molecular biology, bioinformatics, and robotics. Developing tools to democratize molecular biology.
github.com/DominikBuchn...

github.com/DominikBuchn...
Wir betonen 3 besonders wichtige (& einfache) Schritte zur Qualitätssicherung & 4 nächste Schritte (nicht alle einfach).
tinyurl.com/ht-biodiv

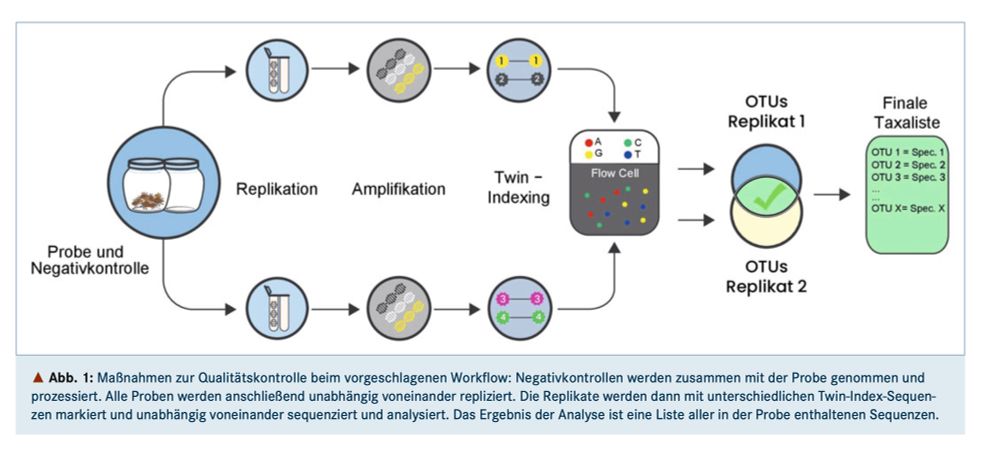

Wir betonen 3 besonders wichtige (& einfache) Schritte zur Qualitätssicherung & 4 nächste Schritte (nicht alle einfach).
tinyurl.com/ht-biodiv
github.com/DominikBuchner…

github.com/DominikBuchner…
protocols.io/view/rna-clean…
protocols.io/view/rna-clean…
I'm desperately searching for small-volume, gravity-flow anion exchange columns comparable to the JetStar columns in the Qiagen RNeasy PowerSoil Total RNA Kit. Does anyone know a supplier for those columns?
I'm desperately searching for small-volume, gravity-flow anion exchange columns comparable to the JetStar columns in the Qiagen RNeasy PowerSoil Total RNA Kit. Does anyone know a supplier for those columns?
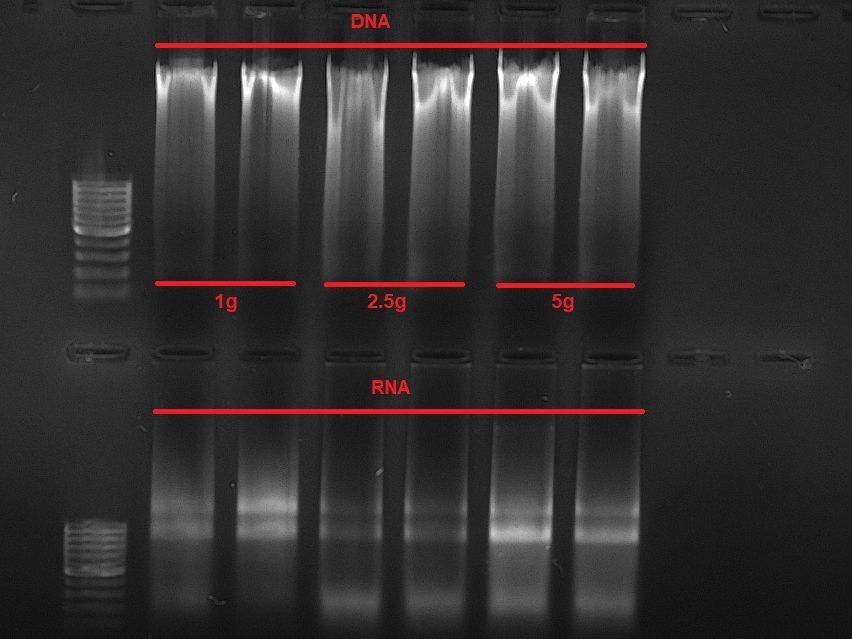


protocols.io/researchers/m4…
protocols.io/researchers/m4…
academic.oup.com/bioinformatics…
academic.oup.com/bioinformatics…


github.com/DominikBuchner…
github.com/DominikBuchner…

sciencedirect.com/science/articl…
sciencedirect.com/science/articl…



mbmg.pensoft.net/article/67533/

mbmg.pensoft.net/article/67533/
onlinelibrary.wiley.com/doi/10.1002/ec…
onlinelibrary.wiley.com/doi/10.1002/ec…



