
Our paper in @naturemethods.bsky.social answers this: use evolutionary history to guide compression and search.
rdcu.be/eg4OA
w/ @baym.lol, @zaminiqbal.bsky.social et al. 🧵1/

Our paper in @naturemethods.bsky.social answers this: use evolutionary history to guide compression and search.
rdcu.be/eg4OA
w/ @baym.lol, @zaminiqbal.bsky.social et al. 🧵1/
Check our latest preprint: doi.org/10.1101/2025...
#Bioinformatics
#Viruses

Check our latest preprint: doi.org/10.1101/2025...
#Bioinformatics
#Viruses
doi.org/10.1093/bioi...
#Bioinformatics #DataCompression

doi.org/10.1093/bioi...
#Bioinformatics #DataCompression
AltaiR: a C toolkit for alignment-free and temporal analysis of multi-FASTA data doi.org/10.1093/giga...
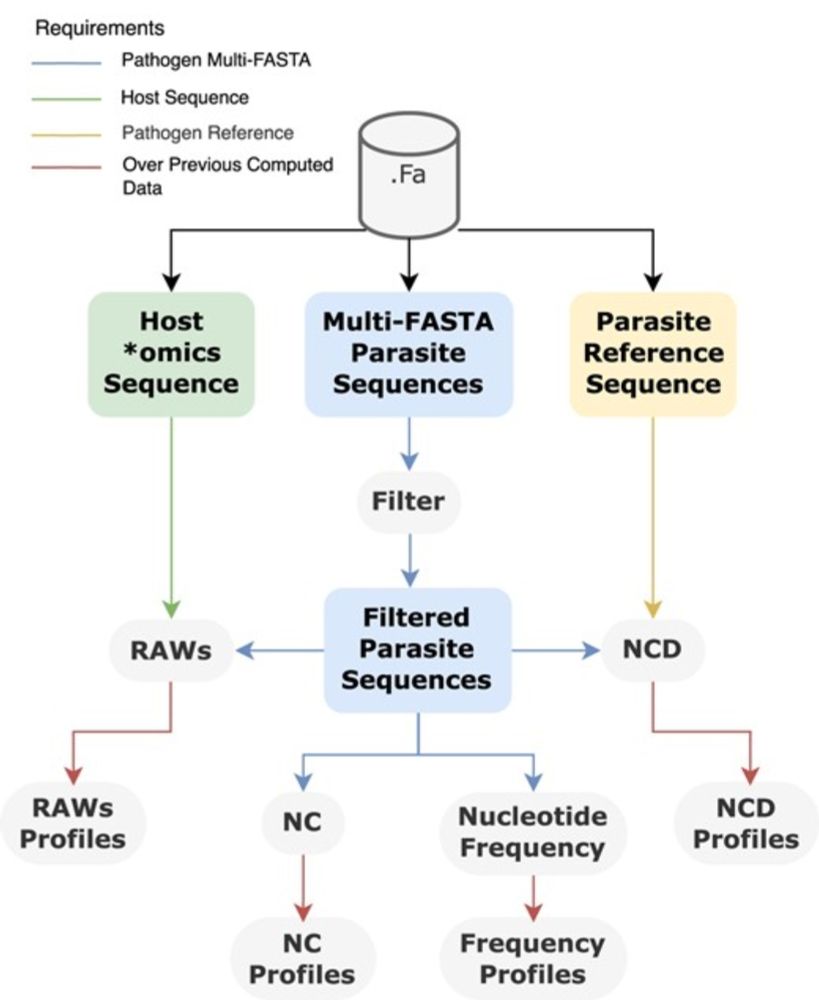
AltaiR: a C toolkit for alignment-free and temporal analysis of multi-FASTA data doi.org/10.1093/giga...
doi.org/10.1093/nar/...

doi.org/10.1093/nar/...
pubs.acs.org/doi/10.1021/...
#Bioremediation #Bioinformatics

pubs.acs.org/doi/10.1021/...
#Bioremediation #Bioinformatics
doi.org/10.1093/giga...
#Bioinformatics #OpenScience

doi.org/10.1093/giga...
#Bioinformatics #OpenScience

