@alandenadel.bsky.social
Reposted
We have recently uploaded our revised manuscript “Evaluating the role of pre-training dataset size and diversity on single-cell foundation model performance”.
TL;DR: More models, more tasks => same results.
TL;DR: More models, more tasks => same results.
November 7, 2025 at 8:07 PM
We have recently uploaded our revised manuscript “Evaluating the role of pre-training dataset size and diversity on single-cell foundation model performance”.
TL;DR: More models, more tasks => same results.
TL;DR: More models, more tasks => same results.
Reposted
Introducing REFLEX, a novel immune profiling assay.
This new method allows for the capture of sequence information for potentially millions of T cells simultaneously.
🔗 https://www.biorxiv.org/content/10.1101/2025.10.24.684243v1
This new method allows for the capture of sequence information for potentially millions of T cells simultaneously.
🔗 https://www.biorxiv.org/content/10.1101/2025.10.24.684243v1
www.biorxiv.org
November 7, 2025 at 11:43 PM
Introducing REFLEX, a novel immune profiling assay.
This new method allows for the capture of sequence information for potentially millions of T cells simultaneously.
🔗 https://www.biorxiv.org/content/10.1101/2025.10.24.684243v1
This new method allows for the capture of sequence information for potentially millions of T cells simultaneously.
🔗 https://www.biorxiv.org/content/10.1101/2025.10.24.684243v1
We have recently uploaded our revised manuscript “Evaluating the role of pre-training dataset size and diversity on single-cell foundation model performance”.
TL;DR: More models, more tasks => same results.
TL;DR: More models, more tasks => same results.
November 7, 2025 at 8:07 PM
We have recently uploaded our revised manuscript “Evaluating the role of pre-training dataset size and diversity on single-cell foundation model performance”.
TL;DR: More models, more tasks => same results.
TL;DR: More models, more tasks => same results.
Reposted
genomebiology.biomedcentral.com/articles/10....
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
Zero-shot evaluation reveals limitations of single-cell foundation models - Genome Biology
Foundation models such as scGPT and Geneformer have not been rigorously evaluated in a setting where they are used without any further training (i.e., zero-shot). Understanding the performance of mode...
genomebiology.biomedcentral.com
April 20, 2025 at 4:14 PM
genomebiology.biomedcentral.com/articles/10....
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
Quite an indictment of some of the current single cell "virtual cell" foundation models. Even for the relatively mundane applications, cell labeling, batch correction etc, they are poor compared to much simpler & cheaper methods.
Reposted
Great to see our paper presenting recall, a framework which calibrates clustering for the impact of data "double-dipping" in single-cell studies, out today in AJHG! Congratulations, @alandenadel.bsky.social and co-authors!
🚨Online now!
📄Artificial variables help to avoid over-clustering in single-cell RNA sequencing
🧑🤝🧑 @alandenadel.bsky.social @lcrawford.bsky.social & co
📄Artificial variables help to avoid over-clustering in single-cell RNA sequencing
🧑🤝🧑 @alandenadel.bsky.social @lcrawford.bsky.social & co

Artificial variables help to avoid over-clustering in single-cell RNA sequencing
Calibrated clustering with artificial variables protects against over-clustering single-cell
RNA-seq data by controlling for the impact of reusing the same data twice when performing
differential expr...
www.cell.com
March 12, 2025 at 7:18 PM
Great to see our paper presenting recall, a framework which calibrates clustering for the impact of data "double-dipping" in single-cell studies, out today in AJHG! Congratulations, @alandenadel.bsky.social and co-authors!
Reposted
🚨Online now!
📄Artificial variables help to avoid over-clustering in single-cell RNA sequencing
🧑🤝🧑 @alandenadel.bsky.social @lcrawford.bsky.social & co
📄Artificial variables help to avoid over-clustering in single-cell RNA sequencing
🧑🤝🧑 @alandenadel.bsky.social @lcrawford.bsky.social & co

Artificial variables help to avoid over-clustering in single-cell RNA sequencing
Calibrated clustering with artificial variables protects against over-clustering single-cell
RNA-seq data by controlling for the impact of reusing the same data twice when performing
differential expr...
www.cell.com
March 12, 2025 at 5:59 PM
🚨Online now!
📄Artificial variables help to avoid over-clustering in single-cell RNA sequencing
🧑🤝🧑 @alandenadel.bsky.social @lcrawford.bsky.social & co
📄Artificial variables help to avoid over-clustering in single-cell RNA sequencing
🧑🤝🧑 @alandenadel.bsky.social @lcrawford.bsky.social & co
Reposted
Can we find epistasis in human traits? To help, in our preprint, we present the most scalable and powerful framework for detecting epistasis to date: the “sparse marginal epistasis test” (SME).
Thank you @lcrawford.bsky.social, @sampatsmith.bsky.social, Dan Weinreich!
doi.org/10.1101/2025...
1/6
Thank you @lcrawford.bsky.social, @sampatsmith.bsky.social, Dan Weinreich!
doi.org/10.1101/2025...
1/6

Sparse modeling of interactions enables fast detection of genome-wide epistasis in biobank-scale studies
The lack of computational methods capable of detecting epistasis in biobanks has led to uncertainty about the role of non-additive genetic effects on complex trait variation. The marginal epistasis fr...
doi.org
January 17, 2025 at 4:46 PM
Can we find epistasis in human traits? To help, in our preprint, we present the most scalable and powerful framework for detecting epistasis to date: the “sparse marginal epistasis test” (SME).
Thank you @lcrawford.bsky.social, @sampatsmith.bsky.social, Dan Weinreich!
doi.org/10.1101/2025...
1/6
Thank you @lcrawford.bsky.social, @sampatsmith.bsky.social, Dan Weinreich!
doi.org/10.1101/2025...
1/6
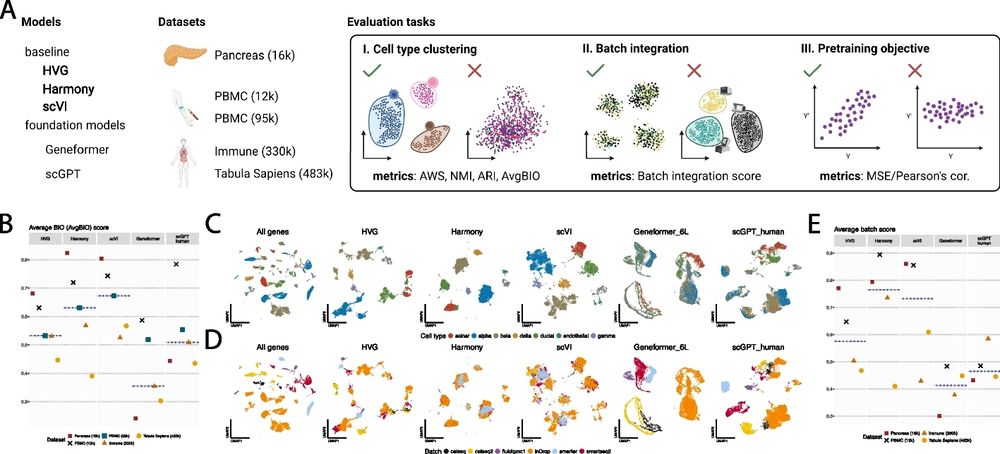
Current methods in the field are trained on atlases ranging from 1 to 100 million cells. In our newest preprint, we show that these same approaches tend to plateau in performance with pre-training datasets that are only a fraction of the size.

December 18, 2024 at 6:48 PM
Current methods in the field are trained on atlases ranging from 1 to 100 million cells. In our newest preprint, we show that these same approaches tend to plateau in performance with pre-training datasets that are only a fraction of the size.

