
Martin Grønbæk-Thygesen (from @rhp-lab.bsky.social) et al show that introducing K here causes ubiquitylation and degradation
doi.org/10.1101/2025...

Martin Grønbæk-Thygesen (from @rhp-lab.bsky.social) et al show that introducing K here causes ubiquitylation and degradation
doi.org/10.1101/2025...
#ChemSky 🧪

#ChemSky 🧪
Tested in some bacterial systems too! Designs so far are short peptides or mini-domains. Looking forward to long/full protein design ahead! 🧪 #microsky
www.nature.com/articles/s41...

Tested in some bacterial systems too! Designs so far are short peptides or mini-domains. Looking forward to long/full protein design ahead! 🧪 #microsky
www.nature.com/articles/s41...
The cover highlights the article on categorizing prediction modes within low-pLDDT regions of AlphaFold2 structures from the Richardson lab @duke-university.bsky.social : doi.org/10.1107/S205...
Image credit: Chistopher Williams

The cover highlights the article on categorizing prediction modes within low-pLDDT regions of AlphaFold2 structures from the Richardson lab @duke-university.bsky.social : doi.org/10.1107/S205...
Image credit: Chistopher Williams
doi.org/10.1101/2025.09.24.678028 #proteindesign

doi.org/10.1101/2025.09.24.678028 #proteindesign
The pLDDT, ipTM, and pTM scores are constantly scoring worse than the same input in AF2, but the PAE is better with AF3.
Anyone else have the same experience?
The pLDDT, ipTM, and pTM scores are constantly scoring worse than the same input in AF2, but the PAE is better with AF3.
Anyone else have the same experience?


Ref.: BioMed Central, 2023
➡️ Continued on ES/IODE
Ref.: BioMed Central, 2023
➡️ Continued on ES/IODE

We compared the protein structure and pathogenicity of clinically relevant variants of the COG4 gene with AlphaFold2 (AF2), Alpha Missense (AM), and…

We compared the protein structure and pathogenicity of clinically relevant variants of the COG4 gene with AlphaFold2 (AF2), Alpha Missense (AM), and…
cellmolbiocommunity.springernature.com/posts/predic...
Pretty cool seeing this (sort of) published. I watched this seminar about it a few months ago and was blown away:
youtu.be/rgGceDDnIEo?...

cellmolbiocommunity.springernature.com/posts/predic...
Pretty cool seeing this (sort of) published. I watched this seminar about it a few months ago and was blown away:
youtu.be/rgGceDDnIEo?...
"Dissecting AlphaFold2’s capabilities with limited sequence information" reveals insights into its ability to predict #ProteinStructures using structural templates without deep MSAs.
Read more: doi.org/10.1093/bioa...
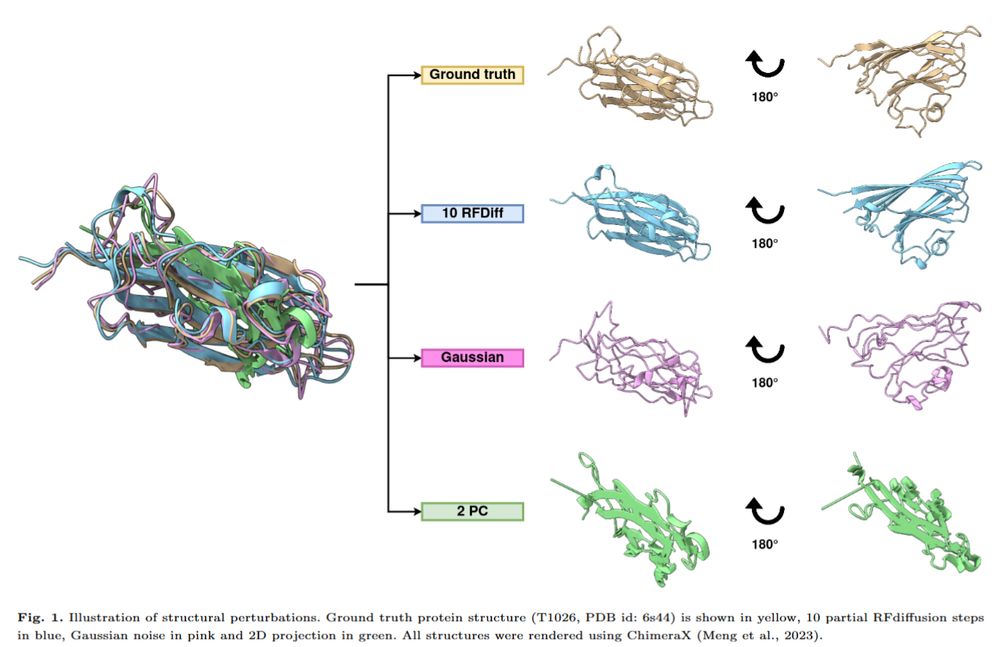
"Dissecting AlphaFold2’s capabilities with limited sequence information" reveals insights into its ability to predict #ProteinStructures using structural templates without deep MSAs.
Read more: doi.org/10.1093/bioa...
www.quantamagazine.org/how-ai-revol...
www.quantamagazine.org/how-ai-revol...

---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
AlphaFold2 refines protein complex models via multimeric templates, enhancing weak contact prediction.




AlphaFold2 refines protein complex models via multimeric templates, enhancing weak contact prediction.
#proteins #structure #prediction #bioinformatics #ISMBECCB2025 #iscbsc

#proteins #structure #prediction #bioinformatics #ISMBECCB2025 #iscbsc
Hand-picked by our Editor-in-Chief as the stand-out article in the issue.
🔦🌱 RESEARCH 🌱🔦
"The nuclear sulfenome of Arabidopsis: spotlight on histone acetyltransferase GCN5 regulation through functional thiols" – De Smet et al. doi.org/10.1093/jxb/...
#PlantScience 🧪

Hand-picked by our Editor-in-Chief as the stand-out article in the issue.
🔦🌱 RESEARCH 🌱🔦
"The nuclear sulfenome of Arabidopsis: spotlight on histone acetyltransferase GCN5 regulation through functional thiols" – De Smet et al. doi.org/10.1093/jxb/...
#PlantScience 🧪


