5/17
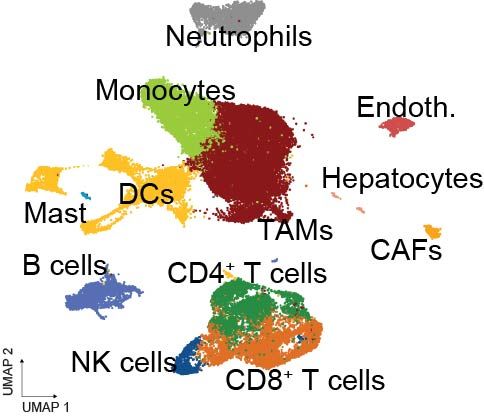
To understand the immune landscape at single-cell resolution, @hoheyn.bsky.social and @paulanietog.bsky.social from @cnag-eu.bsky.social led an extensive scRNA-seq + TCR-seq analysis of tumor-infiltrating lymphocytes (TILs) and myeloid cells in models of metastatic CRC under treatment.
To understand the immune landscape at single-cell resolution, @hoheyn.bsky.social and @paulanietog.bsky.social from @cnag-eu.bsky.social led an extensive scRNA-seq + TCR-seq analysis of tumor-infiltrating lymphocytes (TILs) and myeloid cells in models of metastatic CRC under treatment.
November 7, 2025 at 12:30 PM
5/17
To understand the immune landscape at single-cell resolution, @hoheyn.bsky.social and @paulanietog.bsky.social from @cnag-eu.bsky.social led an extensive scRNA-seq + TCR-seq analysis of tumor-infiltrating lymphocytes (TILs) and myeloid cells in models of metastatic CRC under treatment.
To understand the immune landscape at single-cell resolution, @hoheyn.bsky.social and @paulanietog.bsky.social from @cnag-eu.bsky.social led an extensive scRNA-seq + TCR-seq analysis of tumor-infiltrating lymphocytes (TILs) and myeloid cells in models of metastatic CRC under treatment.
LoFT-TCR: A LoRA-based Fine-tuning Framework for TCR-Antigen Binding Prediction [new]
TCR-antigen binding via LoRA, seq+struct data & graph learning.
TCR-antigen binding via LoRA, seq+struct data & graph learning.




September 27, 2025 at 10:20 PM
LoFT-TCR: A LoRA-based Fine-tuning Framework for TCR-Antigen Binding Prediction [new]
TCR-antigen binding via LoRA, seq+struct data & graph learning.
TCR-antigen binding via LoRA, seq+struct data & graph learning.
🚀 Episode 18 of #OnAIRR is here!
@PGTimmune joins the @airr_community podcast to discuss the evolution of TCR sequencing from single-chain to high-throughput paired TIRTL-seq 🐢
🎧 Listen in any podcast app or here: onairr.airr-community.org
📅 New episodes every month!
#AIRRC
@PGTimmune joins the @airr_community podcast to discuss the evolution of TCR sequencing from single-chain to high-throughput paired TIRTL-seq 🐢
🎧 Listen in any podcast app or here: onairr.airr-community.org
📅 New episodes every month!
#AIRRC

On AIRR - An AIRR Community Podcast - The Antibody Society
The On AIRR podcast series aims to disseminate the goals and values of the Community, with a focus on their application to diagnostics and other clinical applications. We are committed to presenting c...
onairr.airr-community.org
September 25, 2025 at 11:28 AM
🚀 Episode 18 of #OnAIRR is here!
@PGTimmune joins the @airr_community podcast to discuss the evolution of TCR sequencing from single-chain to high-throughput paired TIRTL-seq 🐢
🎧 Listen in any podcast app or here: onairr.airr-community.org
📅 New episodes every month!
#AIRRC
@PGTimmune joins the @airr_community podcast to discuss the evolution of TCR sequencing from single-chain to high-throughput paired TIRTL-seq 🐢
🎧 Listen in any podcast app or here: onairr.airr-community.org
📅 New episodes every month!
#AIRRC
In brief, it:
- Provides the largest spatial data set to date including single cell RNA-seq, TCR profiling and multiplex immunoflourescence
- Reveals three distinct patterns of immune cell composition in chordoma tumors, potentially pointing to a need for treatment strategies tailored for each
- Provides the largest spatial data set to date including single cell RNA-seq, TCR profiling and multiplex immunoflourescence
- Reveals three distinct patterns of immune cell composition in chordoma tumors, potentially pointing to a need for treatment strategies tailored for each
September 22, 2025 at 6:00 PM
In brief, it:
- Provides the largest spatial data set to date including single cell RNA-seq, TCR profiling and multiplex immunoflourescence
- Reveals three distinct patterns of immune cell composition in chordoma tumors, potentially pointing to a need for treatment strategies tailored for each
- Provides the largest spatial data set to date including single cell RNA-seq, TCR profiling and multiplex immunoflourescence
- Reveals three distinct patterns of immune cell composition in chordoma tumors, potentially pointing to a need for treatment strategies tailored for each
23 tools to work with (single-cell) TCR/BCR-seq immune repertoire data 🧵 👇
September 8, 2025 at 2:15 PM
23 tools to work with (single-cell) TCR/BCR-seq immune repertoire data 🧵 👇
RNA-seq based T cell repertoire extraction compared with TCR-seq url: academic.oup.com/ooim/article...

RNA-seq based T cell repertoire extraction compared with TCR-seq
Abstract. The purpose of this study is to evaluate the feasibility of using RNA sequencing data as substrate for the computational extraction of T cell rec
academic.oup.com
August 29, 2025 at 11:51 AM
RNA-seq based T cell repertoire extraction compared with TCR-seq url: academic.oup.com/ooim/article...
Quantifying The Impact of Bulk TCR-Seq Methodological Choices on The Profiled T Cell Repertoire https://www.biorxiv.org/content/10.1101/2025.08.05.668736v1
August 8, 2025 at 12:16 AM
Quantifying The Impact of Bulk TCR-Seq Methodological Choices on The Profiled T Cell Repertoire https://www.biorxiv.org/content/10.1101/2025.08.05.668736v1
Quantifying The Impact of Bulk TCR-Seq Methodological Choices on The Profiled T Cell Repertoire https://www.biorxiv.org/content/10.1101/2025.08.05.668736v1
August 8, 2025 at 12:16 AM
Quantifying The Impact of Bulk TCR-Seq Methodological Choices on The Profiled T Cell Repertoire https://www.biorxiv.org/content/10.1101/2025.08.05.668736v1
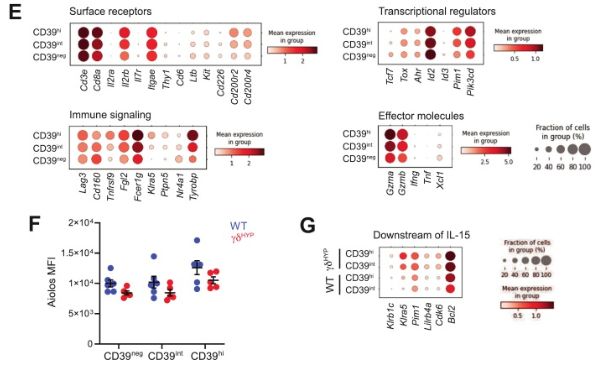
To determine why CD39 expression was increased, we performed CITE-seq and found that CD39 expression positively correlated with transition from a naive-like to tissue-adapted, mature γδ IEL state. Moreover, CD39hi γδ IELs expressed genes downstream of TCR and IL-15 signaling. 4/n
August 7, 2025 at 9:36 PM
To determine why CD39 expression was increased, we performed CITE-seq and found that CD39 expression positively correlated with transition from a naive-like to tissue-adapted, mature γδ IEL state. Moreover, CD39hi γδ IELs expressed genes downstream of TCR and IL-15 signaling. 4/n
New research used single-nucleus RNAseq, spatial transcriptomics, & T cell receptor (TCR) seq. to analyze T cell & glial cell states in post-mortem #Parkinsons brain tissue (cingulate cortex & substantia nigra; N=44)
www.nature.com/articles/s41...
www.nature.com/articles/s41...



August 5, 2025 at 9:41 PM
New research used single-nucleus RNAseq, spatial transcriptomics, & T cell receptor (TCR) seq. to analyze T cell & glial cell states in post-mortem #Parkinsons brain tissue (cingulate cortex & substantia nigra; N=44)
www.nature.com/articles/s41...
www.nature.com/articles/s41...
scRNA + TCR + BCR-seq Reveals the proportion and characteristics of dual TCR T cells and dual BCR B cells in mice infected with Echinococcus granulosus https://www.biorxiv.org/content/10.1101/2025.07.24.666567v1
July 30, 2025 at 5:16 AM
scRNA + TCR + BCR-seq Reveals the proportion and characteristics of dual TCR T cells and dual BCR B cells in mice infected with Echinococcus granulosus https://www.biorxiv.org/content/10.1101/2025.07.24.666567v1
scRNA + TCR + BCR-seq Reveals the proportion and characteristics of dual TCR T cells and dual BCR B cells in mice infected with Echinococcus granulosus https://www.biorxiv.org/content/10.1101/2025.07.24.666567v1
July 30, 2025 at 5:16 AM
scRNA + TCR + BCR-seq Reveals the proportion and characteristics of dual TCR T cells and dual BCR B cells in mice infected with Echinococcus granulosus https://www.biorxiv.org/content/10.1101/2025.07.24.666567v1
Can you statistically test patterns of sparse receptor co- or anti- expression within the cluster? Or must it be approached via CITE-Seq, higher read depth, or sub-library production, as per TCR/BCR repertoire analysis?
June 17, 2025 at 1:30 PM
Can you statistically test patterns of sparse receptor co- or anti- expression within the cluster? Or must it be approached via CITE-Seq, higher read depth, or sub-library production, as per TCR/BCR repertoire analysis?
Groundbreaking #longCOVID study! 🔬 Combining with Live-Seq would let us track immune responses in real-time during epipharyngeal therapy, revealing TCR signaling and cytokine dynamics as they happen. Game-changer for understanding pathophysiology and developing targeted treatments! #SingleCell
June 3, 2025 at 2:08 PM
Groundbreaking #longCOVID study! 🔬 Combining with Live-Seq would let us track immune responses in real-time during epipharyngeal therapy, revealing TCR signaling and cytokine dynamics as they happen. Game-changer for understanding pathophysiology and developing targeted treatments! #SingleCell
1/🧬 This is the first 🧭🧬 spatiotemporal single-cell atlas of human GVHD.
We combined 🧪 Mixed Lymphocyte Reaction, 🧬 TCR tracking, 🔬 single-cell RNA-seq, and 🗺️ spatial transcriptomics to map how donor T cells evolve 🔁, move 🧗♂️, and damage tissue 💥 after transplant.
We combined 🧪 Mixed Lymphocyte Reaction, 🧬 TCR tracking, 🔬 single-cell RNA-seq, and 🗺️ spatial transcriptomics to map how donor T cells evolve 🔁, move 🧗♂️, and damage tissue 💥 after transplant.
May 29, 2025 at 4:48 PM
1/🧬 This is the first 🧭🧬 spatiotemporal single-cell atlas of human GVHD.
We combined 🧪 Mixed Lymphocyte Reaction, 🧬 TCR tracking, 🔬 single-cell RNA-seq, and 🗺️ spatial transcriptomics to map how donor T cells evolve 🔁, move 🧗♂️, and damage tissue 💥 after transplant.
We combined 🧪 Mixed Lymphocyte Reaction, 🧬 TCR tracking, 🔬 single-cell RNA-seq, and 🗺️ spatial transcriptomics to map how donor T cells evolve 🔁, move 🧗♂️, and damage tissue 💥 after transplant.
A new comparative study of commercial single-cell RNAseq kits/chemistries from the Fred Hutchinson Cancer Center: They compared seven whole transcriptome kits and three TCR kits.
scRNA-seq
www.biorxiv.org/content/10.1...
scRNA-seq
www.biorxiv.org/content/10.1...

Evaluating the practical aspects and performance of commercial single-cell RNA sequencing technologies
The rapid development of updated and new commercially available single-cell transcriptomics platforms provides users with a range of experimental options. Cost, sensitivity, throughput, flexibility, a...
www.biorxiv.org
May 27, 2025 at 4:56 PM
A new comparative study of commercial single-cell RNAseq kits/chemistries from the Fred Hutchinson Cancer Center: They compared seven whole transcriptome kits and three TCR kits.
scRNA-seq
www.biorxiv.org/content/10.1...
scRNA-seq
www.biorxiv.org/content/10.1...
Happy to share a new preprint from our group. In studying lung T cell responses in humans, we wondered if the method of sampling matters. Here we isolated paired BAL and endobronchial brushing T cells followed by scRNA- and TCR-seq, finding surprising differences #Immunology
Distinct phenotypes and repertoires of bronchoalveolar and airway mucosal T cells in health and allergic asthma https://www.biorxiv.org/content/10.1101/2025.05.08.652962v1
May 22, 2025 at 3:25 PM
Happy to share a new preprint from our group. In studying lung T cell responses in humans, we wondered if the method of sampling matters. Here we isolated paired BAL and endobronchial brushing T cells followed by scRNA- and TCR-seq, finding surprising differences #Immunology
Tanmana next asked how MEK regulates bioenergetic demand during chronic TCR stimulation. RNA-seq analysis led her to hypothesize that MEK regulates bioenergetic demand in Texh at the level of protein synthesis.

May 14, 2025 at 5:06 PM
Tanmana next asked how MEK regulates bioenergetic demand during chronic TCR stimulation. RNA-seq analysis led her to hypothesize that MEK regulates bioenergetic demand in Texh at the level of protein synthesis.
TCR-seq q: collaborator has ~10% of clones assigned to alleles IMGT says are "ORFs" like TRAJ44*01, TRBJ1-6*01 (this is 10x in mice). What can cause this? How likely are they productive TCRs? Maybe @jamieheather.bsky.social @victorgreiff.bsky.social @10xgenomics.bsky.social have ideas? 🙏
May 9, 2025 at 6:26 PM
TCR-seq q: collaborator has ~10% of clones assigned to alleles IMGT says are "ORFs" like TRAJ44*01, TRBJ1-6*01 (this is 10x in mice). What can cause this? How likely are they productive TCRs? Maybe @jamieheather.bsky.social @victorgreiff.bsky.social @10xgenomics.bsky.social have ideas? 🙏
39 cars entered for 2025 TCR Italy opener at Misano – TouringCarTimes
https://www.byteseu.com/974543/
A massive field of 39 cars have been confirmed for the 2025 TCR Italy season which starts this weekend at Misano, with 20 cars in the main SEQ championship and 19 for the DSG championship which …
https://www.byteseu.com/974543/
A massive field of 39 cars have been confirmed for the 2025 TCR Italy season which starts this weekend at Misano, with 20 cars in the main SEQ championship and 19 for the DSG championship which …

39 cars entered for 2025 TCR Italy opener at Misano – TouringCarTimes - Bytes Europe
A massive field of 39 cars have been confirmed for the 2025 TCR Italy season which starts this weekend at Misano, with 20 cars in the main SEQ championship
www.byteseu.com
May 2, 2025 at 10:26 AM
39 cars entered for 2025 TCR Italy opener at Misano – TouringCarTimes
https://www.byteseu.com/974543/
A massive field of 39 cars have been confirmed for the 2025 TCR Italy season which starts this weekend at Misano, with 20 cars in the main SEQ championship and 19 for the DSG championship which …
https://www.byteseu.com/974543/
A massive field of 39 cars have been confirmed for the 2025 TCR Italy season which starts this weekend at Misano, with 20 cars in the main SEQ championship and 19 for the DSG championship which …
23 tools to work with (single-cell) TCR/BCR-seq immune repertoire data 🧵 👇
April 24, 2025 at 1:45 PM
23 tools to work with (single-cell) TCR/BCR-seq immune repertoire data 🧵 👇
return during the three-year follow-up, compared to seven of the eight who did not respond to the vaccine treatments. Because pancreatic cancer cells are adept at spreading early on, some organs may be harboring undetectable cancer cells that don’t show up on scans, meaning these tumors aren’t




April 16, 2025 at 4:09 PM
return during the three-year follow-up, compared to seven of the eight who did not respond to the vaccine treatments. Because pancreatic cancer cells are adept at spreading early on, some organs may be harboring undetectable cancer cells that don’t show up on scans, meaning these tumors aren’t
Our Services:
📌 Single-cell RNA-seq & ATAC-seq for gene expression & chromatin analysis
📌 Immune profiling (TCR/BCR sequencing)
📌 Single-cell multiomics: combined RNA & chromatin accessibility (ATAC-seq)
📌 Gene expression in fresh, frozen & FFPE samples
📌 Single-cell RNA-seq & ATAC-seq for gene expression & chromatin analysis
📌 Immune profiling (TCR/BCR sequencing)
📌 Single-cell multiomics: combined RNA & chromatin accessibility (ATAC-seq)
📌 Gene expression in fresh, frozen & FFPE samples
April 11, 2025 at 9:27 AM
Our Services:
📌 Single-cell RNA-seq & ATAC-seq for gene expression & chromatin analysis
📌 Immune profiling (TCR/BCR sequencing)
📌 Single-cell multiomics: combined RNA & chromatin accessibility (ATAC-seq)
📌 Gene expression in fresh, frozen & FFPE samples
📌 Single-cell RNA-seq & ATAC-seq for gene expression & chromatin analysis
📌 Immune profiling (TCR/BCR sequencing)
📌 Single-cell multiomics: combined RNA & chromatin accessibility (ATAC-seq)
📌 Gene expression in fresh, frozen & FFPE samples
## 유전자 변이 분석: 단일 세포 RNA 시퀀싱 기반 암 미세환경 내 면역 세포 클론성 분석 연구
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…
## 유전자 변이 분석: 단일 세포 RNA 시퀀싱 기반 암 미세환경 내 면역 세포 클론성 분석 연구
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본 연구에서는 세포 간의 유전자 발현 유사성을 기반으로 클론을 정의하고, 다양한 통계적 기법과 머신러닝 모델을 활용하여 클론의 특성을 분석합니다.
freederia.com
April 2, 2025 at 10:55 PM
## 유전자 변이 분석: 단일 세포 RNA 시퀀싱 기반 암 미세환경 내 면역 세포 클론성 분석 연구
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…
## 유전자 변이 분석: 단일 세포 RNA 시퀀싱 기반 암 미세환경 내 면역 세포 클론성 분석 연구
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…
## 유전자 변이 분석: 단일 세포 RNA 시퀀싱 기반 암 미세환경 내 면역 세포 클론성 분석 연구
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본 연구에서는 세포 간의 유전자 발현 유사성을 기반으로 클론을 정의하고, 다양한 통계적 기법과 머신러닝 모델을 활용하여 클론의 특성을 분석합니다.
freederia.com
April 2, 2025 at 10:55 PM
## 유전자 변이 분석: 단일 세포 RNA 시퀀싱 기반 암 미세환경 내 면역 세포 클론성 분석 연구
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…
**Abstract** 본 연구는 단일 세포 RNA 시퀀싱(scRNA-seq) 데이터를 활용하여 암 미세환경(TME) 내 면역 세포의 클론성(clonality)을 분석하는 새로운 방법론을 제시합니다. 특히, T 세포 수용체(TCR) 시퀀싱 데이터가 없는 경우에도 scRNA-seq 데이터만을 이용하여 TME 내 면역 세포의 클론성을 추론하고, 이를 통해 암 면역 치료의 효율성을 예측할 수 있는 기반을 마련하고자 합니다. 본…

