Looks like yet another case of overhyped results due to poor #bioMLeval evaluation of deep learning models -this time deep docking methods - specifically DiffDock. Look forward to the DiffDock authors response. But dont see any major flaws in this critique. Conclusion is REALLY worth reading!
Been a while since I read a paper like this:
• "What [DiffDock] appears to be doing cannot be considered" docking
• "Results are ... contaminated with near neighbors to test cases"
• "Results for DiffDock were artifactual"
• "Results for other methods were incorrectly done"
arxiv.org/abs/2412.02889
• "What [DiffDock] appears to be doing cannot be considered" docking
• "Results are ... contaminated with near neighbors to test cases"
• "Results for DiffDock were artifactual"
• "Results for other methods were incorrectly done"
arxiv.org/abs/2412.02889

Deep-Learning Based Docking Methods: Fair Comparisons to Conventional Docking Workflows
The diffusion learning method, DiffDock, for docking small-molecule ligands into protein binding sites was recently introduced. Results included comparisons to more conventional docking approaches, wi...
arxiv.org
December 5, 2024 at 11:30 PM
Looks like yet another case of overhyped results due to poor #bioMLeval evaluation of deep learning models -this time deep docking methods - specifically DiffDock. Look forward to the DiffDock authors response. But dont see any major flaws in this critique. Conclusion is REALLY worth reading!
To kick off, for #BioMLeval of Borzoi and the related methods I’ve worked on in collaboration with the Kelley group at Calico, we have used Poisson loss for training, Pearson R and r^2 for validation/test, and causal SNP variant classification of GTEx eQTLs as a separate downstream test
November 22, 2024 at 8:00 PM
To kick off, for #BioMLeval of Borzoi and the related methods I’ve worked on in collaboration with the Kelley group at Calico, we have used Poisson loss for training, Pearson R and r^2 for validation/test, and causal SNP variant classification of GTEx eQTLs as a separate downstream test
It's also worth noting that almost all of the benchmark tasks used in most genomic foundation models papers are really poorly constructed & largely worthless for any real world applications. This paper shows that even on those benchmarks, GFMs can't beat their untrained counterparts. #bioMLeval
February 2, 2025 at 1:19 AM
It's also worth noting that almost all of the benchmark tasks used in most genomic foundation models papers are really poorly constructed & largely worthless for any real world applications. This paper shows that even on those benchmarks, GFMs can't beat their untrained counterparts. #bioMLeval
Yet another story of issues with benchmarks and evaluations in ML4bio + a much stronger and fair benchmark #bioMLeval
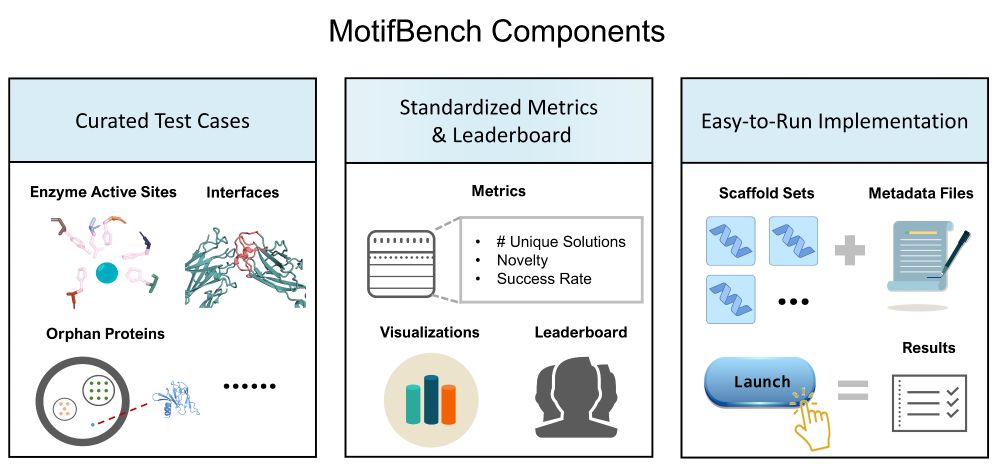
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
February 20, 2025 at 6:00 AM
Yet another story of issues with benchmarks and evaluations in ML4bio + a much stronger and fair benchmark #bioMLeval
Check out this systematic benchmark of genome-wide, annotation agnostic DNALMs & strong baseline ab-initio models for biologically meaningful tasks in regulatory genomics 1/
(1/10) Excited to announce our latest work! @arpita-s.bsky.social, @amanpatel100.bsky.social , and I will be presenting DART-Eval, a rigorous suite of evals for DNA Language Models on transcriptional regulatory DNA at #NeurIPS2024. Check it out! arxiv.org/abs/2412.05430
December 11, 2024 at 3:13 AM
If you're interested in a discussion of statistical methodology for comparison of BioML tools, use the #BioMLeval hashtag. This paper linked by Anshul is a great starting point for discussion. It's oriented towards LLMs, but some ideas may be transferable. What benchmarks do you use for BioML?
www.anthropic.com/research/sta...
This is an excellent attempt (blog & paper) at bringing more statistical rigor to evaluation of ML models (this is specifically focused on LLM evals).
I feel like we need to have similar clear standards for many types of predictive models in biology. 1/
This is an excellent attempt (blog & paper) at bringing more statistical rigor to evaluation of ML models (this is specifically focused on LLM evals).
I feel like we need to have similar clear standards for many types of predictive models in biology. 1/

A statistical approach to model evaluations
A research paper from Anthropic on how to apply statistics to improve language model evaluations
www.anthropic.com
November 22, 2024 at 7:31 PM
If you're interested in a discussion of statistical methodology for comparison of BioML tools, use the #BioMLeval hashtag. This paper linked by Anshul is a great starting point for discussion. It's oriented towards LLMs, but some ideas may be transferable. What benchmarks do you use for BioML?
Also want to point to this other recent preprint that also shows that optimized ab-initio CNN models beat DNALMs even on the (relatively pointless) surrogate tasks used in the DNALM papers. CNNs also beat several other foundation models in other domains.
arxiv.org/abs/2411.02796
#bioMLeval
arxiv.org/abs/2411.02796
#bioMLeval
December 12, 2024 at 6:14 AM
Also want to point to this other recent preprint that also shows that optimized ab-initio CNN models beat DNALMs even on the (relatively pointless) surrogate tasks used in the DNALM papers. CNNs also beat several other foundation models in other domains.
arxiv.org/abs/2411.02796
#bioMLeval
arxiv.org/abs/2411.02796
#bioMLeval
Alright let’s try this. I think we have to first post at least one skeet with the hashtag, e.g. #BioMLeval …
November 22, 2024 at 7:20 PM
Alright let’s try this. I think we have to first post at least one skeet with the hashtag, e.g. #BioMLeval …
I saw the discussion on #BioMLeval pop up thanks to this post and @ianholmes.org. I think this is an interesting + extremely valuable discussion - super happy to see people interested in bioML eval.
www.anthropic.com/research/sta...
This is an excellent attempt (blog & paper) at bringing more statistical rigor to evaluation of ML models (this is specifically focused on LLM evals).
I feel like we need to have similar clear standards for many types of predictive models in biology. 1/
This is an excellent attempt (blog & paper) at bringing more statistical rigor to evaluation of ML models (this is specifically focused on LLM evals).
I feel like we need to have similar clear standards for many types of predictive models in biology. 1/

A statistical approach to model evaluations
A research paper from Anthropic on how to apply statistics to improve language model evaluations
www.anthropic.com
November 23, 2024 at 6:00 PM
I saw the discussion on #BioMLeval pop up thanks to this post and @ianholmes.org. I think this is an interesting + extremely valuable discussion - super happy to see people interested in bioML eval.
Very well-informed thread on #BioMLeval
Such a framework shouldn’t impose guidelines on users, only provide a convenient way to run all kinds of tests on a research prototype (so different from the models folks use in clinic applications).
November 23, 2024 at 6:55 PM
Very well-informed thread on #BioMLeval
This started with @avsecz.bsky.social’s Enformer (I’ve basically just inherited scripts from Borzoi that did the SNP tests). I’ve seen lots of other cool #BioMLeval benchmarks for downstream tasks, like predicting RNA half-lives and ribosome loads in @bowang87.bsky.social’s recent Orthrus paper
November 22, 2024 at 8:04 PM
This started with @avsecz.bsky.social’s Enformer (I’ve basically just inherited scripts from Borzoi that did the SNP tests). I’ve seen lots of other cool #BioMLeval benchmarks for downstream tasks, like predicting RNA half-lives and ribosome loads in @bowang87.bsky.social’s recent Orthrus paper
I think it’s ongoing, on the #BioMLeval hashtag, at a low-key level so far. I may post some more stuff there later
November 24, 2024 at 2:40 AM
I think it’s ongoing, on the #BioMLeval hashtag, at a low-key level so far. I may post some more stuff there later
What a surprise (not!). Yet again ... poor evaluations of specialized medical LLMs result in overhyped claims relative to the base LLMs. #bioMLeval
Medically adapted foundation models (think Med-*) turn out to be more hot air than hot stuff. Correcting for fatal flaws in evaluation, the current crop are no better on balance than generic foundation models, even on the very tasks for which benefits are claimed.
arxiv.org/abs/2411.04118
arxiv.org/abs/2411.04118

Medical Adaptation of Large Language and Vision-Language Models: Are We Making Progress?
Several recent works seek to develop foundation models specifically for medical applications, adapting general-purpose large language models (LLMs) and vision-language models (VLMs) via continued pret...
arxiv.org
November 27, 2024 at 2:16 AM
What a surprise (not!). Yet again ... poor evaluations of specialized medical LLMs result in overhyped claims relative to the base LLMs. #bioMLeval
OK, so now #BioMLeval posts are available via search bsky.app/hashtag/BioM... ... to actually build a custom feed apparently requires setting up a server: docs.bsky.app/docs/starter... ... but maybe search is sufficient for now
bsky.app
November 22, 2024 at 7:25 PM
OK, so now #BioMLeval posts are available via search bsky.app/hashtag/BioM... ... to actually build a custom feed apparently requires setting up a server: docs.bsky.app/docs/starter... ... but maybe search is sufficient for now

