You can extract DNA region modules & visualize CIRCE's interactions over gene locations.
CIRCE also facilitates #CellOracle or #HuMMuS usage, making it unnecessary to run Cicero (code in R) first.
4/5

You can extract DNA region modules & visualize CIRCE's interactions over gene locations.
CIRCE also facilitates #CellOracle or #HuMMuS usage, making it unnecessary to run Cicero (code in R) first.
4/5
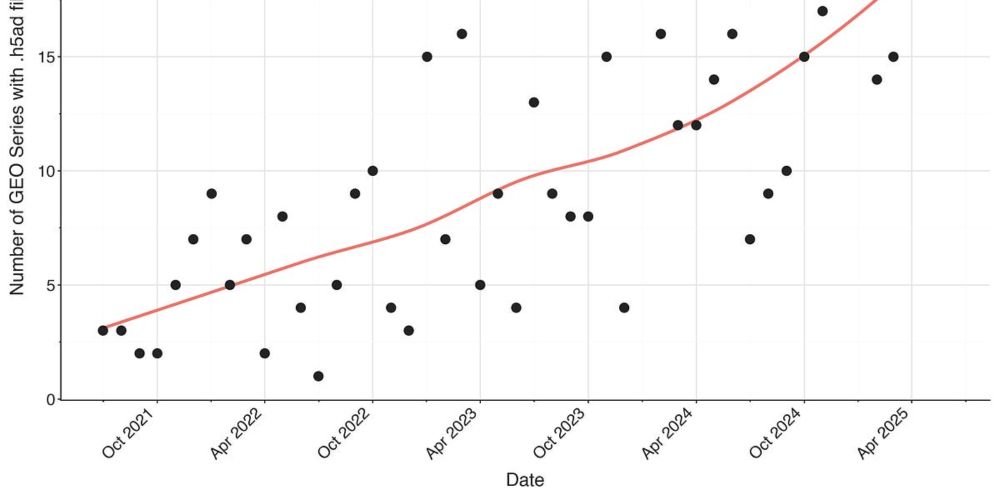
The new .h5sc format provides fast random access to single-cell images for ML training.
It follows #FAIR data principles (findable, accessible, interoperable, reusable) and integrates with @scverse.bsky.social tools via AnnData.

The new .h5sc format provides fast random access to single-cell images for ML training.
It follows #FAIR data principles (findable, accessible, interoperable, reusable) and integrates with @scverse.bsky.social tools via AnnData.

- Read and write H5AD files natively in R
- Convert between AnnData, SingleCellExperiment & Seurat
- Work with AnnData objects in R
If you've struggled with reading H5AD files in R, or single cell format conversion, try out anndataR! anndatar.data-intuitive.com
- Read and write H5AD files natively in R
- Convert between AnnData, SingleCellExperiment & Seurat
- Work with AnnData objects in R
If you've struggled with reading H5AD files in R, or single cell format conversion, try out anndataR! anndatar.data-intuitive.com
🔗 Read the paper: www.biorxiv.org/content/10.1...
💻 Check the package in action: anndatar.data-intuitive.com

🔗 Read the paper: www.biorxiv.org/content/10.1...
💻 Check the package in action: anndatar.data-intuitive.com

AnnData can easily be read into R/Bioc as well unlocking more stats tools.
AnnData can easily be read into R/Bioc as well unlocking more stats tools.
(Zoom registration link and more information in thread!)
(Zoom registration link and more information in thread!)
(Zoom registration link and more information in thread!)
(Zoom registration link and more information in thread!)
- Works seamlessly with Scanpy, Squidpy and AnnData (@scverse_team compatible)
- Plug-and-play any ST dataset
- Outputs spatially informed scores
- Geostatistics-inspired metrics to evaluate gene sets
- Works seamlessly with Scanpy, Squidpy and AnnData (@scverse_team compatible)
- Plug-and-play any ST dataset
- Outputs spatially informed scores
- Geostatistics-inspired metrics to evaluate gene sets
github.com/honicky/annd...
It uses partial downloads to dramatically speed up extracting the metadata without downloading the whole file.
`pip install anndata-metadata`

github.com/honicky/annd...
It uses partial downloads to dramatically speed up extracting the metadata without downloading the whole file.
`pip install anndata-metadata`
What started as a shared vision for interoperable single-cell analysis has become a vibrant, global community.
From AnnData to full multimodal pipelines, we’re building the future of everything single-cell and spatial omics, together.
Here’s to what’s next!
What started as a shared vision for interoperable single-cell analysis has become a vibrant, global community.
From AnnData to full multimodal pipelines, we’re building the future of everything single-cell and spatial omics, together.
Here’s to what’s next!
#Bioinformatics #SingleCell #DuckDb
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf105
Authors: @kennypavan.com and @your-arpy.bsky.social

#Bioinformatics #SingleCell #DuckDb
We hope it helps researchers build flexible, powerful analysis pipelines for analyzing highly multiplexed fluorescence images.
🔧 Package: github.com/sagar87/spat...

We hope it helps researchers build flexible, powerful analysis pipelines for analyzing highly multiplexed fluorescence images.
🔧 Package: github.com/sagar87/spat...