
Willie Neiswanger
@willieneis.bsky.social
Assistant Professor in CS + AI at USC. Previously at Stanford, CMU. Machine Learning, Decision Making, AI-for-Science, Generative AI, ML Systems, LLMs.
https://willieneis.github.io
https://willieneis.github.io
Our paper also contains an in-depth discussion on safety when releasing metagenomic models.
Looking for collaborators to build on this with us — please reach out!
metagene.ai
Looking for collaborators to build on this with us — please reach out!
metagene.ai
January 7, 2025 at 8:58 PM
Our paper also contains an in-depth discussion on safety when releasing metagenomic models.
Looking for collaborators to build on this with us — please reach out!
metagene.ai
Looking for collaborators to build on this with us — please reach out!
metagene.ai
We leverage the ecosystem of modern LLM tooling—in tokenization, model architecture, training, infra, etc—for performance and extensibility. METAGENE-1 is standardized & easy to use.
Hugging Face: huggingface.co/metagene-ai
Github: github.com/metagene-ai
Hugging Face: huggingface.co/metagene-ai
Github: github.com/metagene-ai
January 7, 2025 at 8:58 PM
We leverage the ecosystem of modern LLM tooling—in tokenization, model architecture, training, infra, etc—for performance and extensibility. METAGENE-1 is standardized & easy to use.
Hugging Face: huggingface.co/metagene-ai
Github: github.com/metagene-ai
Hugging Face: huggingface.co/metagene-ai
Github: github.com/metagene-ai
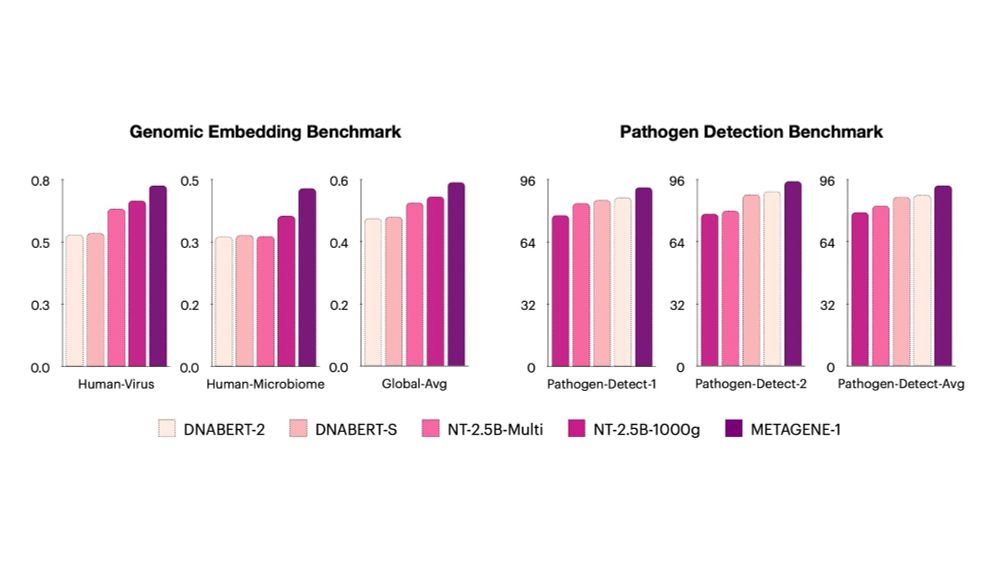
METAGENE-1 shows state-of-the-art results on pathogen detection, metagenomic embedding, and other genomic tasks.
We also release new benchmarks for genomic detection and embedding (eg, Gene-MTEB, based on MTEB for LLMs).
See our paper for details: arxiv.org/abs/2501.02045
We also release new benchmarks for genomic detection and embedding (eg, Gene-MTEB, based on MTEB for LLMs).
See our paper for details: arxiv.org/abs/2501.02045
January 7, 2025 at 8:58 PM
METAGENE-1 shows state-of-the-art results on pathogen detection, metagenomic embedding, and other genomic tasks.
We also release new benchmarks for genomic detection and embedding (eg, Gene-MTEB, based on MTEB for LLMs).
See our paper for details: arxiv.org/abs/2501.02045
We also release new benchmarks for genomic detection and embedding (eg, Gene-MTEB, based on MTEB for LLMs).
See our paper for details: arxiv.org/abs/2501.02045
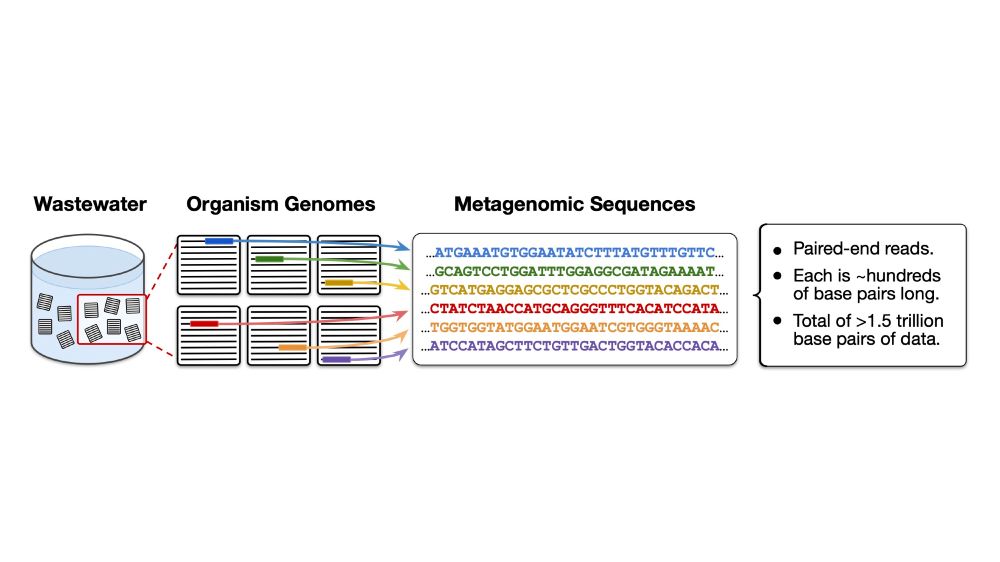
Our data pipeline is: human microbiome > wastewater > metagenomic sequences > tokens > training data.
Wastewater provides a rich source of data from tens of thousands of species across the human-adjacent microbiome. In total we pretrain on over 1.5T base pairs of DNA/RNA.
Wastewater provides a rich source of data from tens of thousands of species across the human-adjacent microbiome. In total we pretrain on over 1.5T base pairs of DNA/RNA.
January 7, 2025 at 8:58 PM
Our data pipeline is: human microbiome > wastewater > metagenomic sequences > tokens > training data.
Wastewater provides a rich source of data from tens of thousands of species across the human-adjacent microbiome. In total we pretrain on over 1.5T base pairs of DNA/RNA.
Wastewater provides a rich source of data from tens of thousands of species across the human-adjacent microbiome. In total we pretrain on over 1.5T base pairs of DNA/RNA.
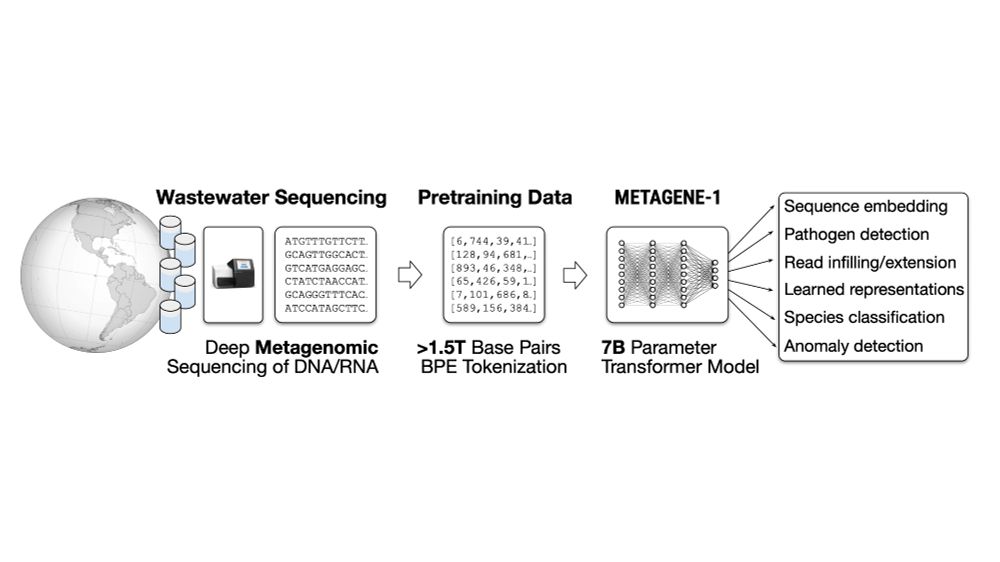
Metagenomic sequencing of wastewater produces vast amounts of data that can capture public health trends at a societal scale. Our goal is to train a model on this data to help in large-scale wastewater monitoring & detection of novel bio threats.
January 7, 2025 at 8:58 PM
Metagenomic sequencing of wastewater produces vast amounts of data that can capture public health trends at a societal scale. Our goal is to train a model on this data to help in large-scale wastewater monitoring & detection of novel bio threats.

