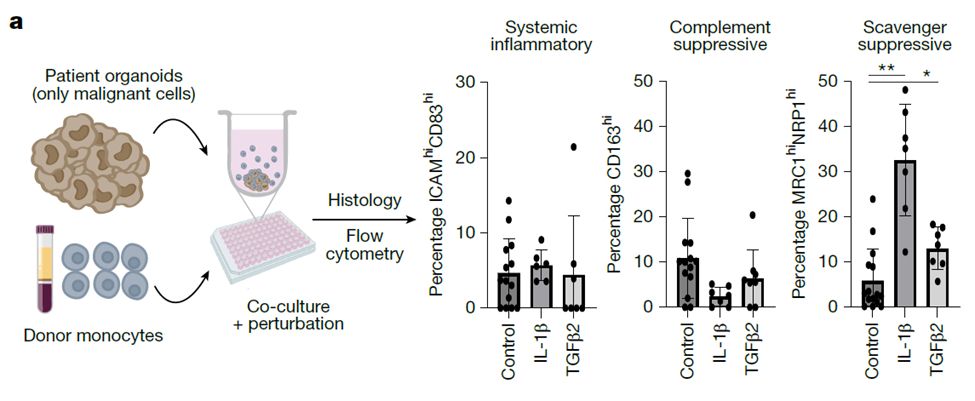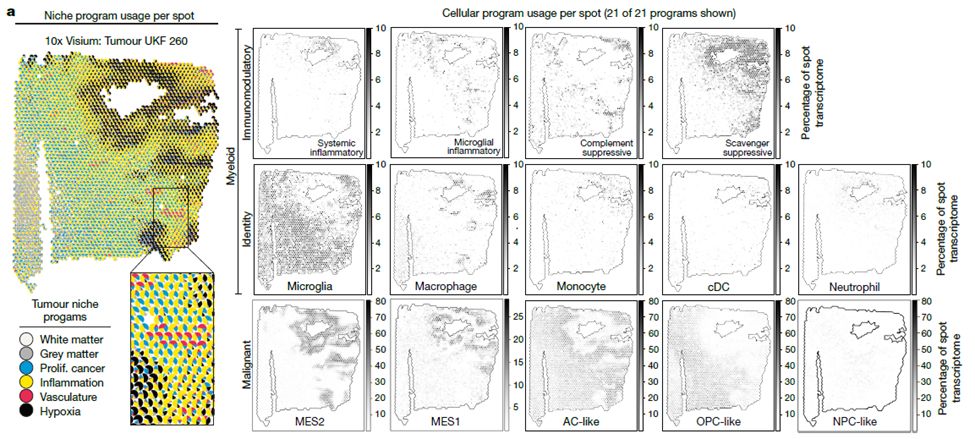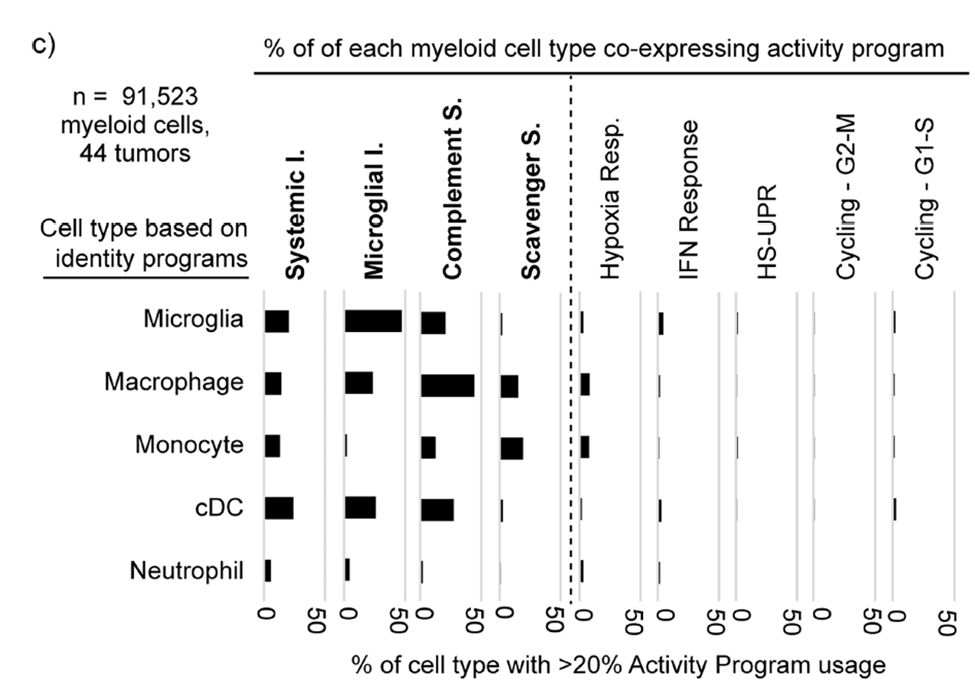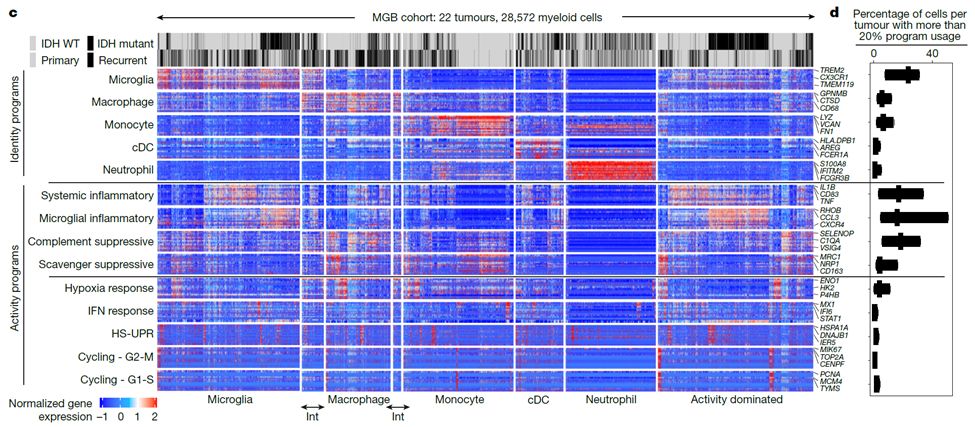
Grateful to have @vrishab_x's mother visit the lab from Seattle and cook us all authentic South Indian food for lunch today 😋❤️

Grateful to have @vrishab_x's mother visit the lab from Seattle and cook us all authentic South Indian food for lunch today 😋❤️

Please share positions below:

Please share positions below: