Tong Wang
@twangmdphd.bsky.social
Resident physician at Stanford Pathology | Greenleaf Lab | Penn MSTP | Interested in chemical biology, epigenetics, and clinically useful tests.
Pinned
Tong Wang
@twangmdphd.bsky.social
· Sep 29

1/ Happy to share our preprint from the Greenleaf Lab: beCasKAS, our method to directly detect CRISPR base editor off-targets in primary cells. We additionally show how non-coding edits can be triaged for epigenetic dysregulation using deep learning.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Tong Wang
After leukemia treatments failed, Alyssa Tapley, at age 13, was told she would die. Then, doctors tried an experimental gene-edited therapy. She became the first human to try the treatment made possible by U.S. federal funding. Now 16, she’s cancer-free and planning her future.
November 10, 2025 at 12:33 AM
After leukemia treatments failed, Alyssa Tapley, at age 13, was told she would die. Then, doctors tried an experimental gene-edited therapy. She became the first human to try the treatment made possible by U.S. federal funding. Now 16, she’s cancer-free and planning her future.
Reposted by Tong Wang
Very well done story on 60 Minutes about the harms of grant terminations and freezes at Harvard with Joan Brugge, Don Engbar, David Liu, and a very compelling young cancer patient, now cured with Liu's technology.
Transcript and video here
www.cbsnews.com/news/researc...
Transcript and video here
www.cbsnews.com/news/researc...

Battle between Trump and universities hurting scientific research in need of federal funding
Federal research funding to universities has fueled breakthroughs for years. The White House is pressuring universities to align with the president's political agenda, or risk losing their funding.
www.cbsnews.com
November 10, 2025 at 1:03 AM
Very well done story on 60 Minutes about the harms of grant terminations and freezes at Harvard with Joan Brugge, Don Engbar, David Liu, and a very compelling young cancer patient, now cured with Liu's technology.
Transcript and video here
www.cbsnews.com/news/researc...
Transcript and video here
www.cbsnews.com/news/researc...
Reposted by Tong Wang
The Editor Got a Letter From ‘Dr. B.S.’ So Did a Lot of Other Editors.
The rise of artificial intelligence has produced serial writers to science and medical journals, most likely using chatbots to boost the number of citations they’ve published
#GiftLink
www.nytimes.com/2025/11/04/s...
The rise of artificial intelligence has produced serial writers to science and medical journals, most likely using chatbots to boost the number of citations they’ve published
#GiftLink
www.nytimes.com/2025/11/04/s...

The Editor Got a Letter From ‘Dr. B.S.’ So Did a Lot of Other Editors.
www.nytimes.com
November 7, 2025 at 10:21 PM
The Editor Got a Letter From ‘Dr. B.S.’ So Did a Lot of Other Editors.
The rise of artificial intelligence has produced serial writers to science and medical journals, most likely using chatbots to boost the number of citations they’ve published
#GiftLink
www.nytimes.com/2025/11/04/s...
The rise of artificial intelligence has produced serial writers to science and medical journals, most likely using chatbots to boost the number of citations they’ve published
#GiftLink
www.nytimes.com/2025/11/04/s...
Reposted by Tong Wang
We then used sequence-to-activity deep learning models, to predict effects of non-coding edits on TF binding and chromatin accessibility. We first show that a ChromBPNet model can predict the same GATA site disruption mechanism exploited by the FDA-approved Casgevy medicine, specifically in T cells:

November 7, 2025 at 6:38 PM
We then used sequence-to-activity deep learning models, to predict effects of non-coding edits on TF binding and chromatin accessibility. We first show that a ChromBPNet model can predict the same GATA site disruption mechanism exploited by the FDA-approved Casgevy medicine, specifically in T cells:
Reposted by Tong Wang
A highlight of my summer was this collab w/
@twangmdphd.bsky.social ! CRISPR base editors are driving several clinical trials, but it's hard to quantify off-target edits and their effects. Tong developed a sequencing assay to identify off-targets *in primary cells* 🧬🖥️🧪 #genomics #chromatin #CRISPR
@twangmdphd.bsky.social ! CRISPR base editors are driving several clinical trials, but it's hard to quantify off-target edits and their effects. Tong developed a sequencing assay to identify off-targets *in primary cells* 🧬🖥️🧪 #genomics #chromatin #CRISPR
1/ Happy to share our preprint from the Greenleaf Lab: beCasKAS, our method to directly detect CRISPR base editor off-targets in primary cells. We additionally show how non-coding edits can be triaged for epigenetic dysregulation using deep learning.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 7, 2025 at 6:38 PM
A highlight of my summer was this collab w/
@twangmdphd.bsky.social ! CRISPR base editors are driving several clinical trials, but it's hard to quantify off-target edits and their effects. Tong developed a sequencing assay to identify off-targets *in primary cells* 🧬🖥️🧪 #genomics #chromatin #CRISPR
@twangmdphd.bsky.social ! CRISPR base editors are driving several clinical trials, but it's hard to quantify off-target edits and their effects. Tong developed a sequencing assay to identify off-targets *in primary cells* 🧬🖥️🧪 #genomics #chromatin #CRISPR
Reposted by Tong Wang
Sensitive, direct detection of non-coding off-target base editor unwinding and editing in primary cells [new]
Direct seq. quantifies off-target base editor & DNA unwinding.
Direct seq. quantifies off-target base editor & DNA unwinding.




September 26, 2025 at 4:55 AM
Sensitive, direct detection of non-coding off-target base editor unwinding and editing in primary cells [new]
Direct seq. quantifies off-target base editor & DNA unwinding.
Direct seq. quantifies off-target base editor & DNA unwinding.
Reposted by Tong Wang
Sensitive, direct detection of non-coding off-target base editor unwinding and editing in primary cells https://www.biorxiv.org/content/10.1101/2025.09.25.678665v1
September 26, 2025 at 4:33 AM
Sensitive, direct detection of non-coding off-target base editor unwinding and editing in primary cells https://www.biorxiv.org/content/10.1101/2025.09.25.678665v1
1/ Happy to share our preprint from the Greenleaf Lab: beCasKAS, our method to directly detect CRISPR base editor off-targets in primary cells. We additionally show how non-coding edits can be triaged for epigenetic dysregulation using deep learning.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

September 29, 2025 at 6:28 PM
1/ Happy to share our preprint from the Greenleaf Lab: beCasKAS, our method to directly detect CRISPR base editor off-targets in primary cells. We additionally show how non-coding edits can be triaged for epigenetic dysregulation using deep learning.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Tong Wang
Delighted to share our latest work deciphering the landscape of chromatin accessibility and modeling the DNA sequence syntax rules underlying gene regulation during human fetal development! www.biorxiv.org/content/10.1... Read on for more: 🧵 1/16 #GeneReg 🧬🖥️

Dissecting regulatory syntax in human development with scalable multiomics and deep learning
Transcription factors (TFs) establish cell identity during development by binding regulatory DNA in a sequence-specific manner, often promoting local chromatin accessibility, and regulating gene expre...
www.biorxiv.org
May 3, 2025 at 6:27 PM
Delighted to share our latest work deciphering the landscape of chromatin accessibility and modeling the DNA sequence syntax rules underlying gene regulation during human fetal development! www.biorxiv.org/content/10.1... Read on for more: 🧵 1/16 #GeneReg 🧬🖥️
Reposted by Tong Wang
www.biorxiv.org/content/10.1...
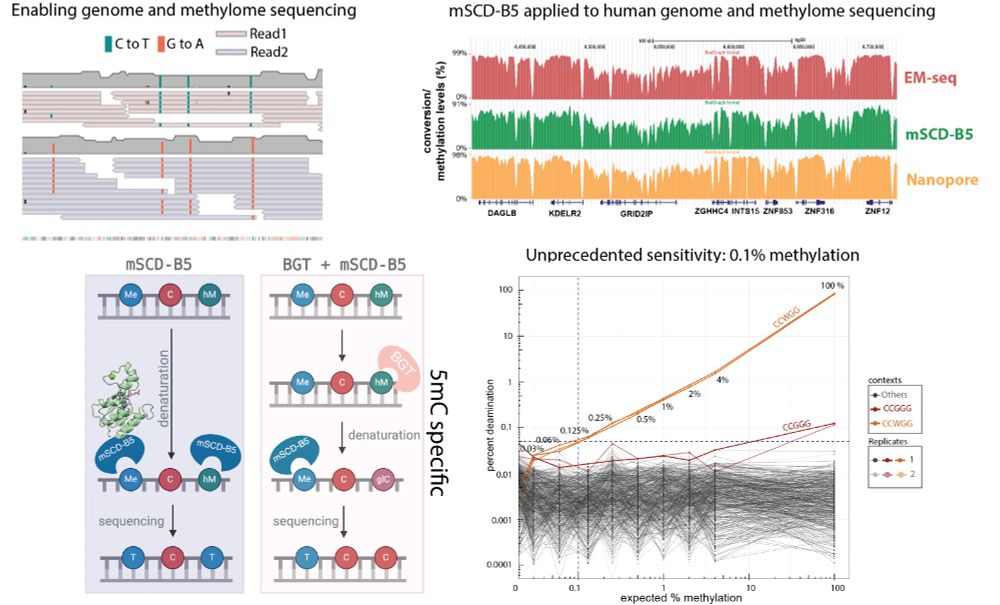
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
December 9, 2024 at 2:12 PM
www.biorxiv.org/content/10.1...
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
Our study published today on #bioRxiv describe the identification of a deaminase that converts 5mC to T, enabling direct sequencing of the human methylome and genome. This achievement was made possible through a collaborative effort across all departments at #NEB.
Reposted by Tong Wang
(1st post @BlueSky) Preprint alert🚨a long thread. Cautions in the use of @nanopore sequencing to map DNA modifications: officially reported “accuracy” ≠ reliable mapping in real applications. We performed a critical assessment of nanopore sequencing (across different versions of models) for the 1/n

November 20, 2024 at 11:41 AM
(1st post @BlueSky) Preprint alert🚨a long thread. Cautions in the use of @nanopore sequencing to map DNA modifications: officially reported “accuracy” ≠ reliable mapping in real applications. We performed a critical assessment of nanopore sequencing (across different versions of models) for the 1/n
Reposted by Tong Wang
who knew it would be easier to get everyone to switch social media platforms than getting them to use hg38
November 14, 2024 at 6:45 PM
who knew it would be easier to get everyone to switch social media platforms than getting them to use hg38

