
And a special thank you to the talented @mfernandezquintero.bsky.social & Johannes Loeffler for the stunning graphic! 🎨
#CancerResearch #Immunotherapy #AI
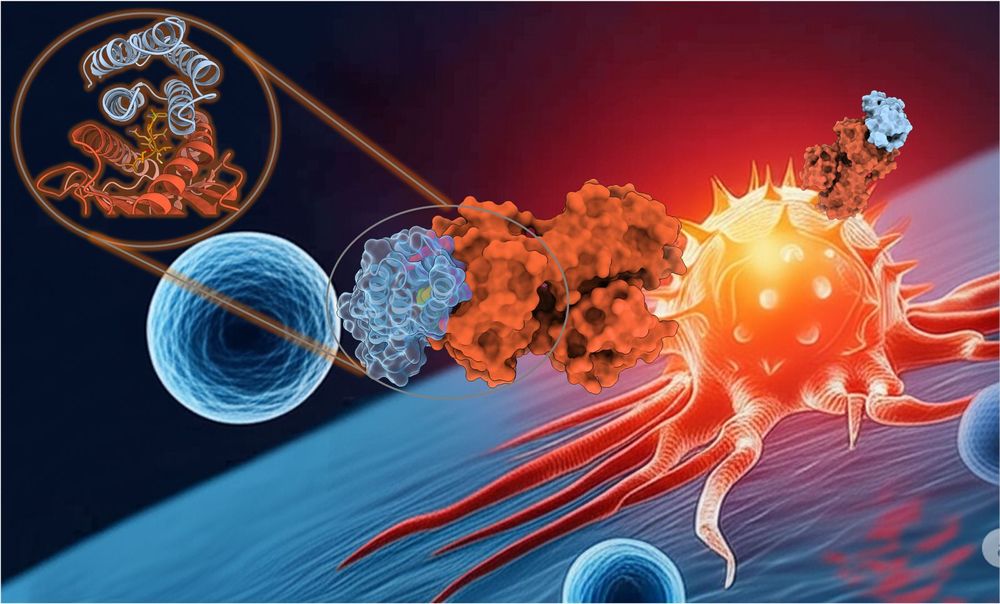
And a special thank you to the talented @mfernandezquintero.bsky.social & Johannes Loeffler for the stunning graphic! 🎨
#CancerResearch #Immunotherapy #AI
A huge congratulations to the incredible teams at DTU HealthTech, DTU Bioengineering, & @scripps.edu and to all my brilliant co-authors, especially @Kristoffer Haurum Johansen.

A huge congratulations to the incredible teams at DTU HealthTech, DTU Bioengineering, & @scripps.edu and to all my brilliant co-authors, especially @Kristoffer Haurum Johansen.
⚡️ Speed: A rapid 4-6 week pipeline from computer design to validated lab result.
🎯 Precision: An AI-driven process to screen for specificity & reduce off-target risks.
🤝 Personalization: It works even for a patient's unique tumor mutation.

⚡️ Speed: A rapid 4-6 week pipeline from computer design to validated lab result.
🎯 Precision: An AI-driven process to screen for specificity & reduce off-target risks.
🤝 Personalization: It works even for a patient's unique tumor mutation.
These act as a high-precision guidance system, leading a patient's own T-cells (black) to recognize and destroy cancer cells (red) with remarkable accuracy.
#ProteinDesign #GenerativeAI
These act as a high-precision guidance system, leading a patient's own T-cells (black) to recognize and destroy cancer cells (red) with remarkable accuracy.
#ProteinDesign #GenerativeAI
– How generative AI is transforming protein research?
– Why mass spec is such a rich (but messy) playground for AI?
– What it takes to go from spectrum to sequence to structure?
Give it a listen 🎧
– How generative AI is transforming protein research?
– Why mass spec is such a rich (but messy) playground for AI?
– What it takes to go from spectrum to sequence to structure?
Give it a listen 🎧
📄 www.nature.com/articles/s42...
📄 www.nature.com/articles/s42...
– Proteins that are hard to purify
– Assays that demand specificity
– Targets that need quantitative detection
Please reach out if you want to hear more.
#StartupLaunch #Founder #AffinityAI

– Proteins that are hard to purify
– Assays that demand specificity
– Targets that need quantitative detection
Please reach out if you want to hear more.
#StartupLaunch #Founder #AffinityAI
– Designerbodies™ with pM–nM affinity
– ARCANE™ designs for affinity, stability, and expression
– RAVEN™ screens candidates at scale
Built for diagnostics, therapeutics, and biotech pipelines.
#AIinBiotech #SyntheticBiology

– Designerbodies™ with pM–nM affinity
– ARCANE™ designs for affinity, stability, and expression
– RAVEN™ screens candidates at scale
Built for diagnostics, therapeutics, and biotech pipelines.
#AIinBiotech #SyntheticBiology
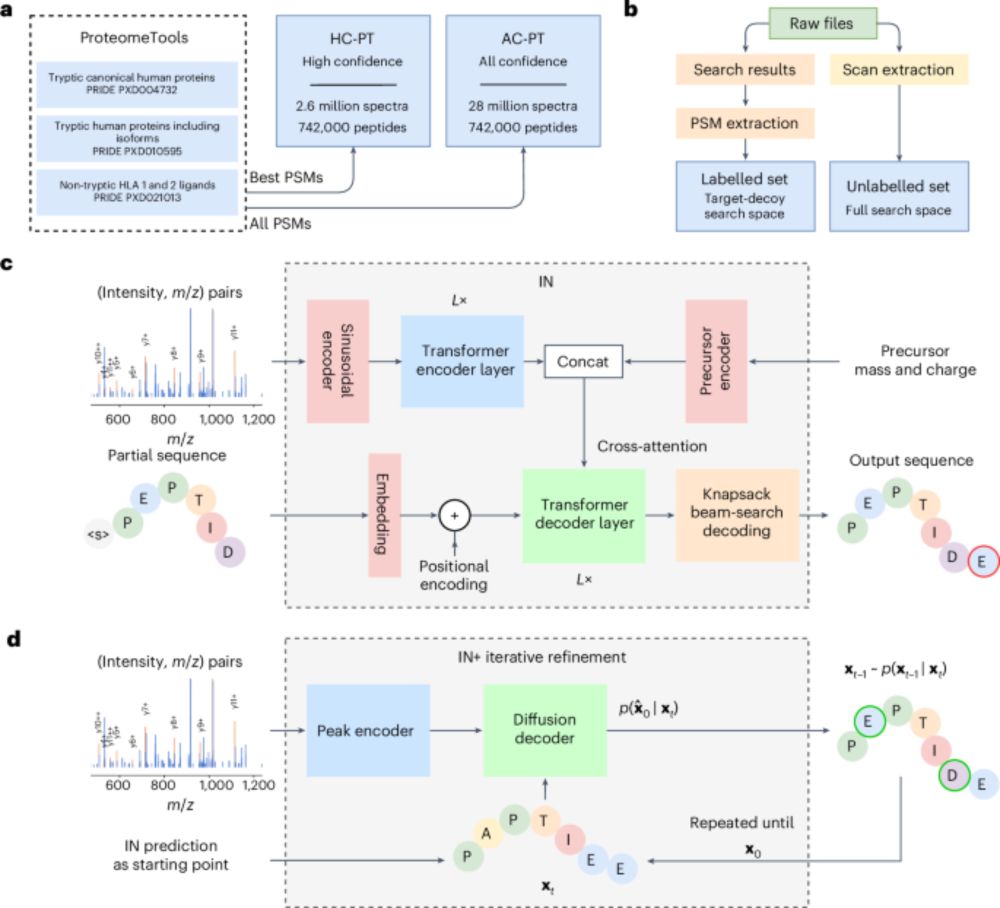
v1.1 generalises well:
In a GluC-treated HeLa dataset, we reached 81.3% peptide recall—higher depth, same model.
Improved confidence metrics = better precision, fewer false positives, more IDs.
Code: bit.ly/43ExRGI
v1.1 generalises well:
In a GluC-treated HeLa dataset, we reached 81.3% peptide recall—higher depth, same model.
Improved confidence metrics = better precision, fewer false positives, more IDs.
Code: bit.ly/43ExRGI
What’s new in InstaNovo v1.1?
🧬 +13.5% recall
🎯 +42.6% more exact PSMs
🔍 145% more peptides & 35% more proteins @ 5% FDR
🧪 Support for 7 PTMs: phosphorylation, deamidation, carbamylation, ammonia loss, oxidation, acetylation, carbamidomethylation
Paper: bit.ly/3XBeJFh
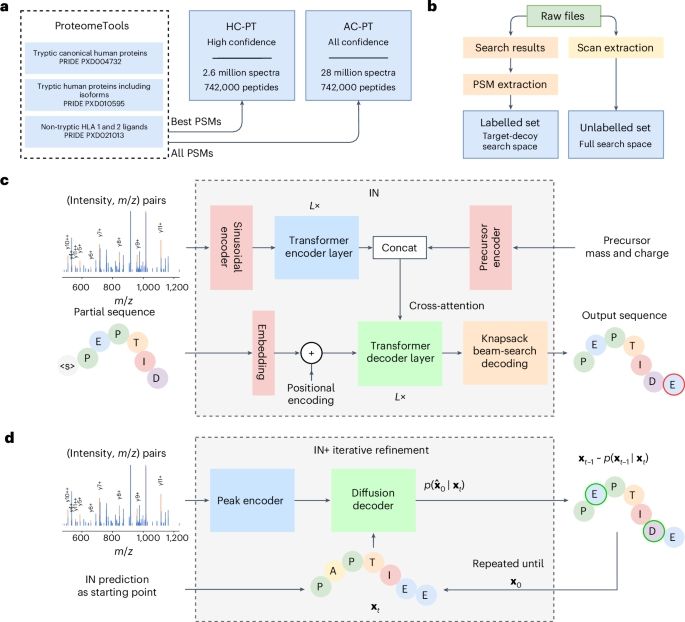
What’s new in InstaNovo v1.1?
🧬 +13.5% recall
🎯 +42.6% more exact PSMs
🔍 145% more peptides & 35% more proteins @ 5% FDR
🧪 Support for 7 PTMs: phosphorylation, deamidation, carbamylation, ammonia loss, oxidation, acetylation, carbamidomethylation
Paper: bit.ly/3XBeJFh

