
🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...

🌐 afesm.foldseek.com
📄 www.biorxiv.org/content/10.1...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
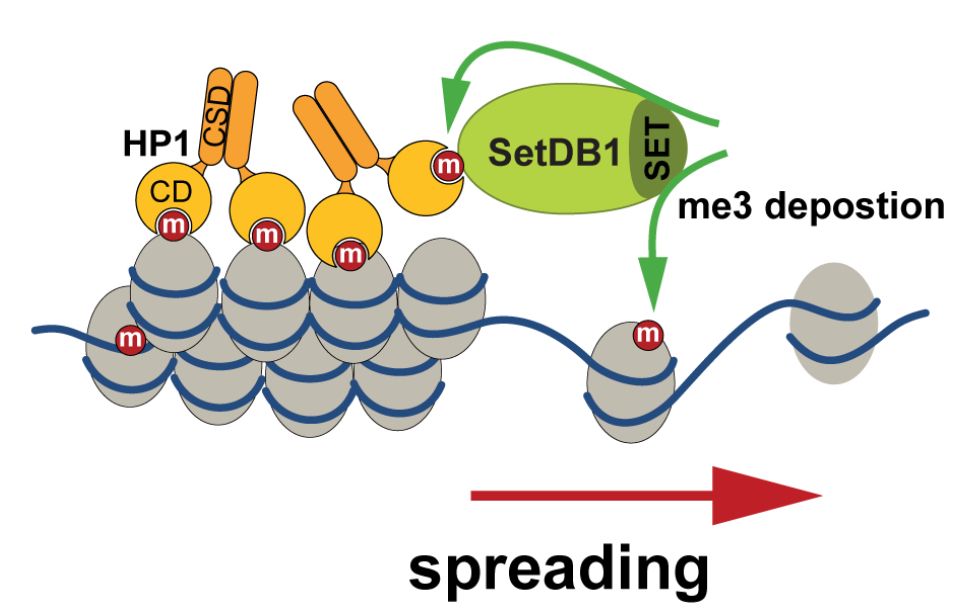
@masaashimazoe.bsky.social et al. reveal that linker histone H1 acts as a liquid-like glue to organize chromatin in living cells. 🎉 Fantastic collab with @rcollepardo.bsky.social @janhuemar.bsky.social and others—huge thanks! 🙌 1/
www.ks.uiuc.edu/Research/vmd...

www.ks.uiuc.edu/Research/vmd...
To me, the most important are:
Read often, read broadly (incl. older papers and outside your field), and learn to read some papers in detail and others more superficially (and quickly)

To me, the most important are:
Read often, read broadly (incl. older papers and outside your field), and learn to read some papers in detail and others more superficially (and quickly)
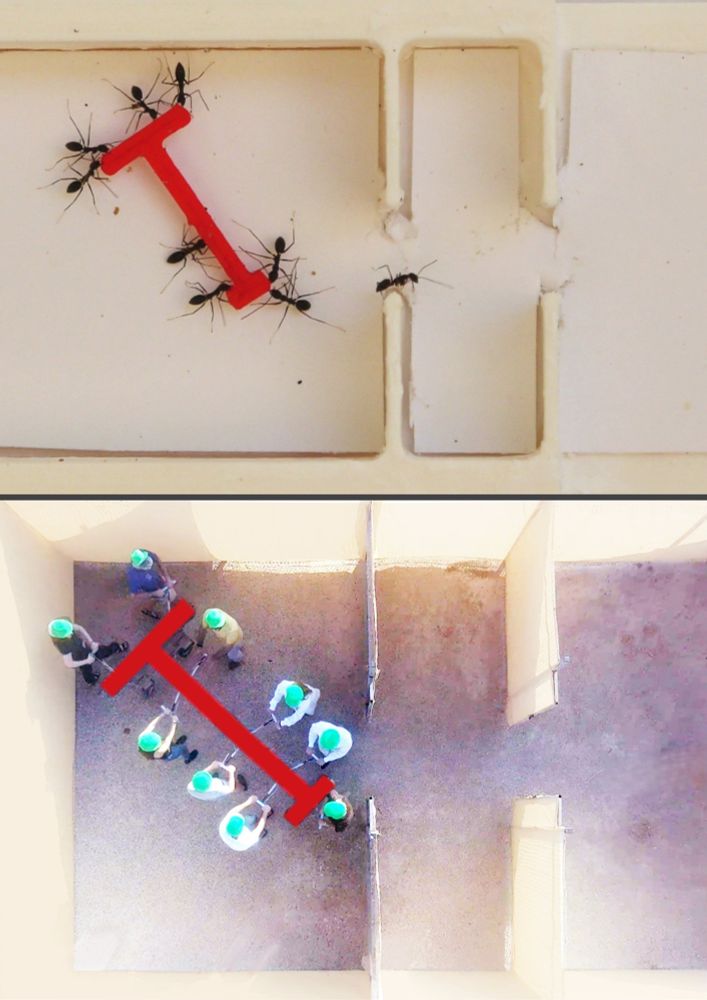
We address this at near-atomistic resolution🔥🔥🔥 using cryoET (Rosen & Villa labs, led by H Zhou), a new multiscale model (K Russell) and cryoET-guided sims (J Huertas & J Maristany)
We address this at near-atomistic resolution🔥🔥🔥 using cryoET (Rosen & Villa labs, led by H Zhou), a new multiscale model (K Russell) and cryoET-guided sims (J Huertas & J Maristany)


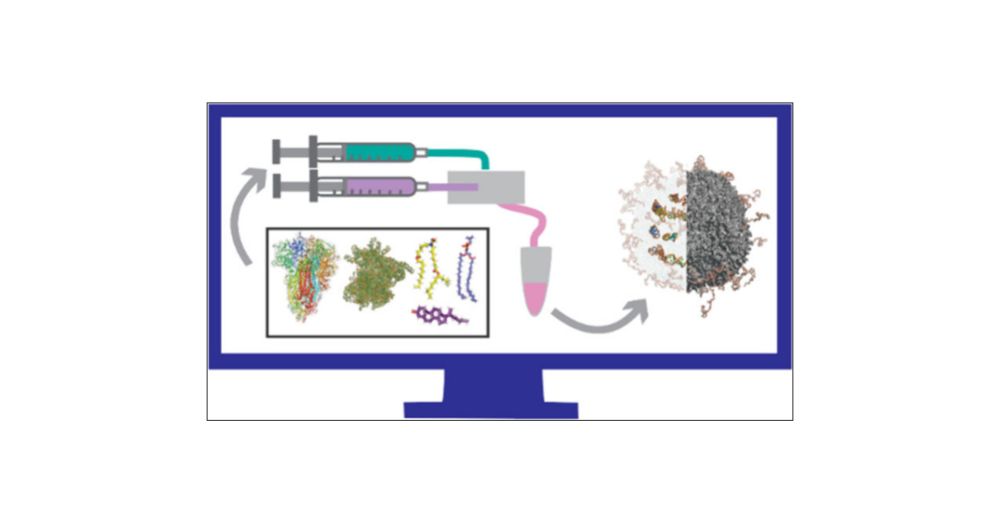
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

From @commsbio.bsky.social's "Reliability and reproducibility checklist for molecular dynamics simulations" (doi.org/10.1038/s420...)
IMO the number 3 is meaningless and could equally well be 1 or 1000
From @commsbio.bsky.social's "Reliability and reproducibility checklist for molecular dynamics simulations" (doi.org/10.1038/s420...)
IMO the number 3 is meaningless and could equally well be 1 or 1000
graph-tool.skewed.de
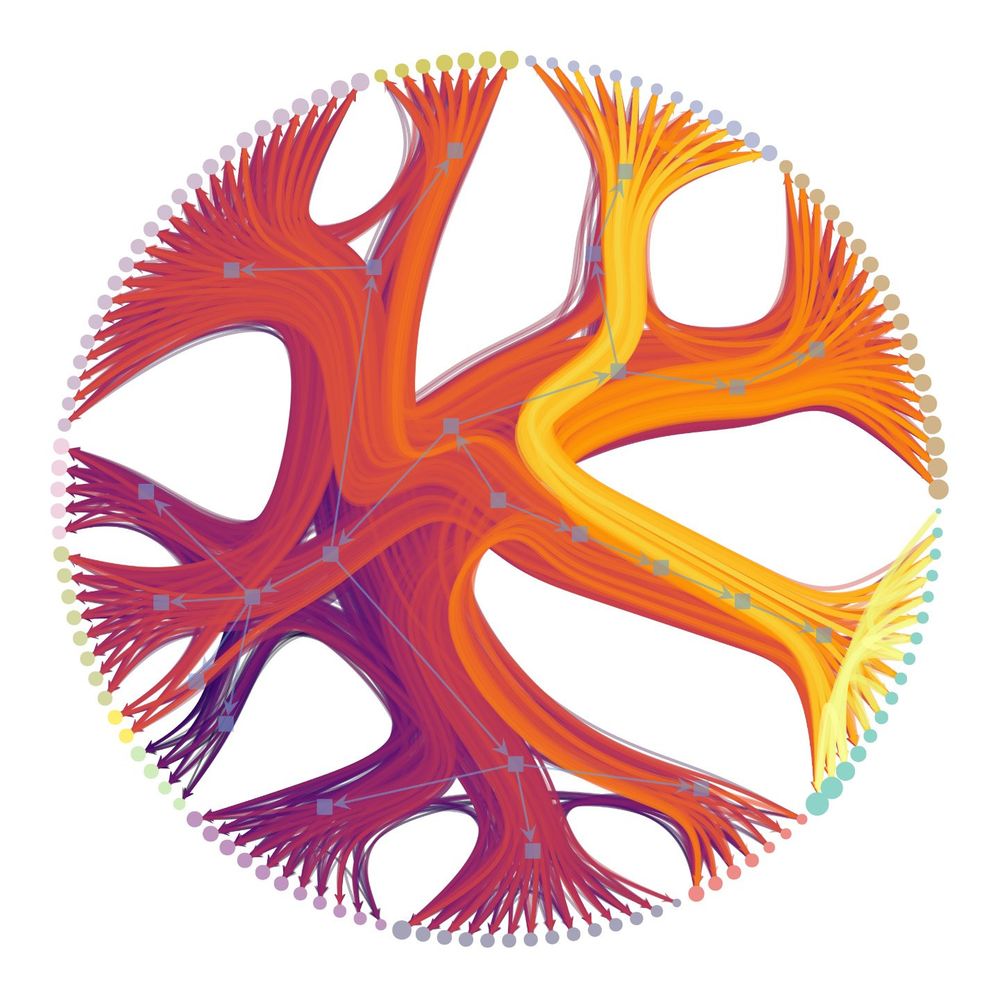
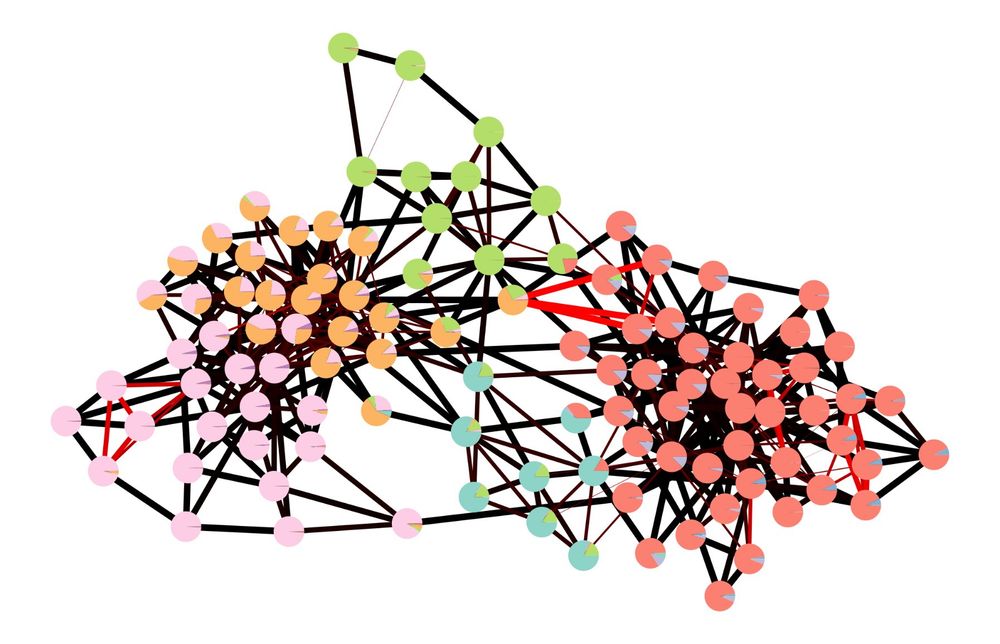
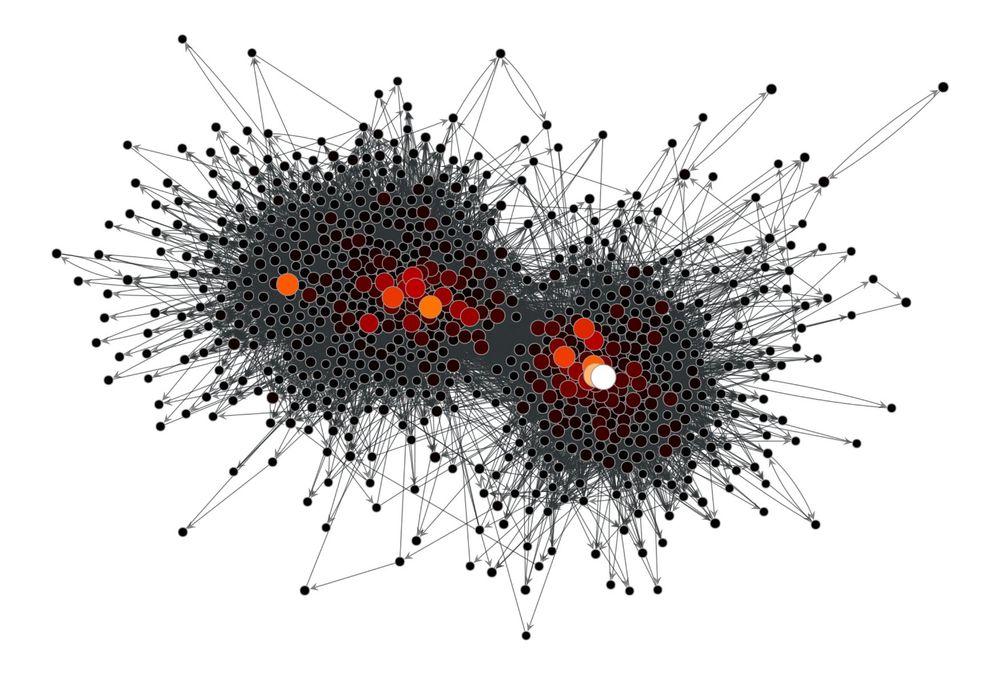
Graph-tool is a comprehensive and efficient Python library to work with networks, including structural, dynamical, and statistical algorithms, as well as visualization. 1/N
#networkscience
graph-tool.skewed.de
Graph-tool is a comprehensive and efficient Python library to work with networks, including structural, dynamical, and statistical algorithms, as well as visualization. 1/N
#networkscience
www.biophysics.org/profiles/kre...

www.biophysics.org/profiles/kre...
@philosophaki.bsky.social
Insights into ryanodine receptor activation & calcium-induced Ca2+ release from a stochastic explicit-particle 3D simulation of cardiac dyad w/ realistic TEM geometry 🧪
www.sciencedirect.com/science/arti...
@philosophaki.bsky.social
Insights into ryanodine receptor activation & calcium-induced Ca2+ release from a stochastic explicit-particle 3D simulation of cardiac dyad w/ realistic TEM geometry 🧪
www.sciencedirect.com/science/arti...
pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!

pubs.acs.org/doi/10.1021/...
@pabloarantes.bsky.social & team also made it into a Colab notebook:
colab.research.google.com/drive/1GGJKr...
Give it a try & send feedback!

