suragnair.github.io


They allow parallel decoding, with competitive performance at fewer generation steps. 8/


They allow parallel decoding, with competitive performance at fewer generation steps. 8/







Nona operates on both DNA sequence and functional genomics tracks. Task-specific masking configurations recover familiar model types, and its flexibility enables entirely new approaches! 2/
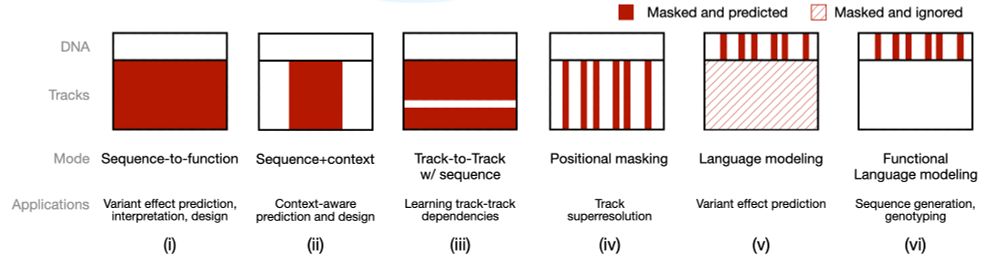
Nona operates on both DNA sequence and functional genomics tracks. Task-specific masking configurations recover familiar model types, and its flexibility enables entirely new approaches! 2/
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15

Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15

