
Mingyuan "Merlin" Li
@supmerlin.bsky.social
PhD candidate at Johns Hopkins | Alexis Battle lab | Computational Biology | Machine Learning | He/Him
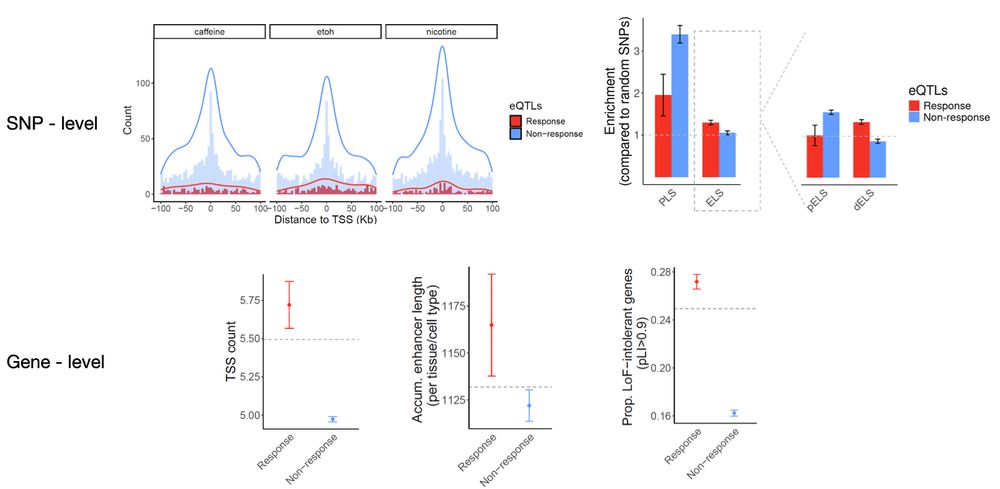
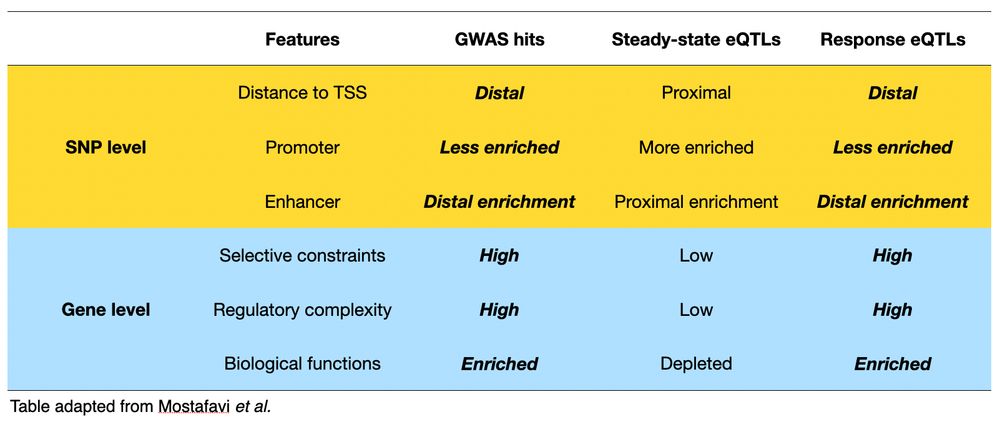
Thanks for the question! I think this result is consistent with the previous result (figure 3) where we found response eQTLs share more similar genomic features with disease-associated locus.
May 27, 2025 at 3:24 PM
Thanks for the question! I think this result is consistent with the previous result (figure 3) where we found response eQTLs share more similar genomic features with disease-associated locus.
This is such an exciting project! Huge thanks to Wenhe Lin, @jmpopp.bsky.social, @radster95.bsky.social, Olivia Allen, Jonathan Burnett, and to @alexisbattle.bsky.social, @ygilad.bsky.social, and Matthew Stephens for their mentorships! Check out our preprint for more!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

May 5, 2025 at 7:07 PM
This is such an exciting project! Huge thanks to Wenhe Lin, @jmpopp.bsky.social, @radster95.bsky.social, Olivia Allen, Jonathan Burnett, and to @alexisbattle.bsky.social, @ygilad.bsky.social, and Matthew Stephens for their mentorships! Check out our preprint for more!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
9/n We expect that by continuing to broaden the contexts to include a wider range of cell types, time points, and environmental exposures, we will uncover more context-dependent and functionally relevant loci, which will enhance our ability to explain disease risk.🌐🧬
May 5, 2025 at 7:07 PM
9/n We expect that by continuing to broaden the contexts to include a wider range of cell types, time points, and environmental exposures, we will uncover more context-dependent and functionally relevant loci, which will enhance our ability to explain disease risk.🌐🧬
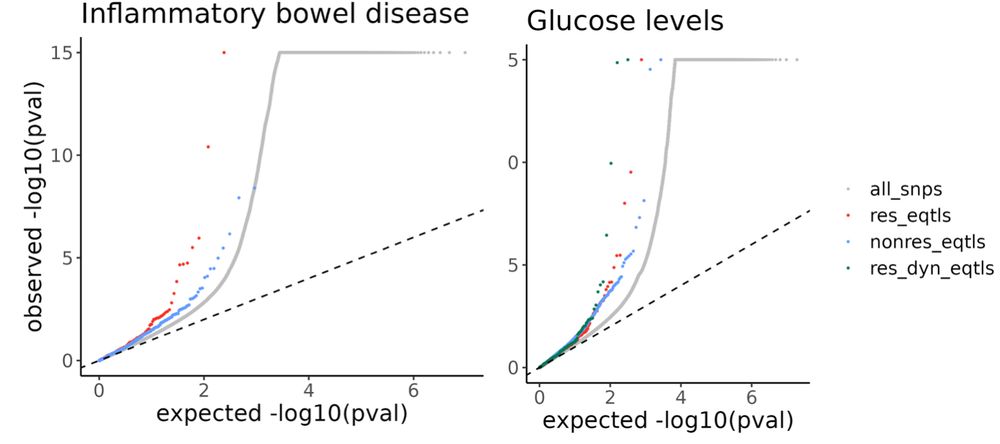
8/n We found that incorporating more context-specificity enabled us to identify regulatory effects that may enhance the functional understanding of GWAS associations. These effects would have been missed had we only explored a single tissue type or not accounted for GxE.
May 5, 2025 at 7:07 PM
8/n We found that incorporating more context-specificity enabled us to identify regulatory effects that may enhance the functional understanding of GWAS associations. These effects would have been missed had we only explored a single tissue type or not accounted for GxE.
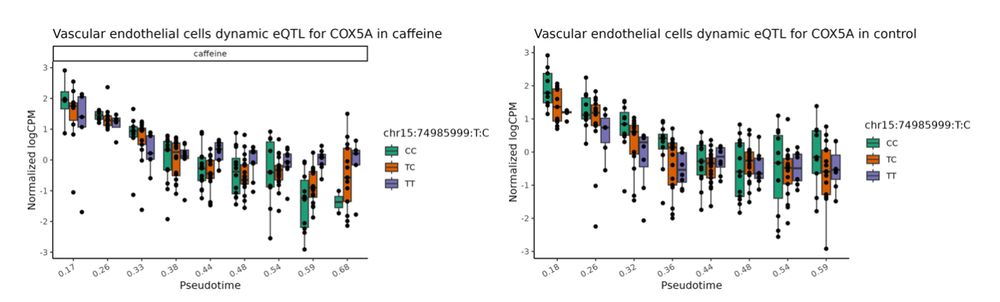
7/n We also mapped dynamic response eQTLs that vary their regulatory effects throughout cellular differentiation. Example: A caffeine-response dynamic eQTL affecting COX5A emerged exclusively in late-stage vascular differentiation. ☕️🩸✨
May 5, 2025 at 7:07 PM
7/n We also mapped dynamic response eQTLs that vary their regulatory effects throughout cellular differentiation. Example: A caffeine-response dynamic eQTL affecting COX5A emerged exclusively in late-stage vascular differentiation. ☕️🩸✨
6/n Importantly, we found that response eQTLs share important properties with disease-associated GWAS loci, including similar regulatory landscapes, gene-level constraint, and functional enrichments.📍🔍
May 5, 2025 at 7:07 PM
6/n Importantly, we found that response eQTLs share important properties with disease-associated GWAS loci, including similar regulatory landscapes, gene-level constraint, and functional enrichments.📍🔍
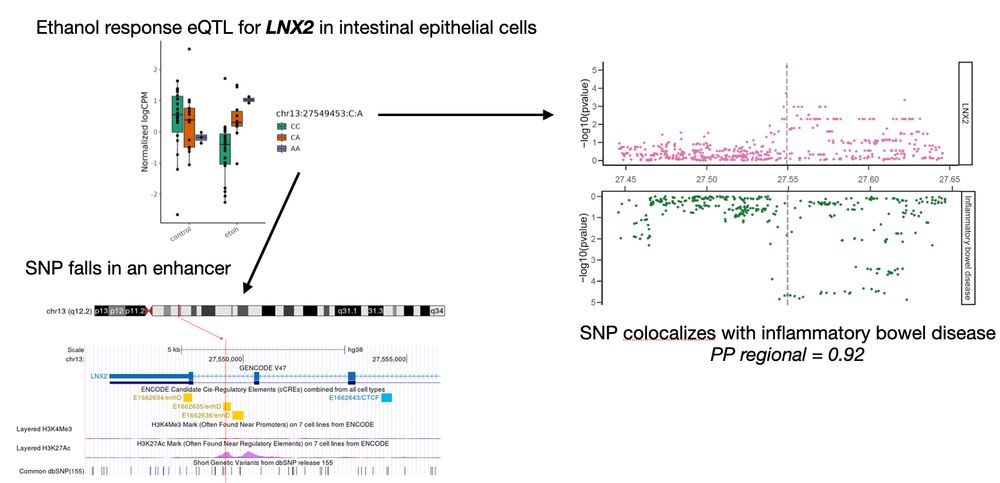
5/n suggesting a functional mechanistic connection between a gene regulatory variant, ethanol exposure, LNX2 expression in the intestinal epithelium, and IBD. 🍷➡️🩺
May 5, 2025 at 7:07 PM
5/n suggesting a functional mechanistic connection between a gene regulatory variant, ethanol exposure, LNX2 expression in the intestinal epithelium, and IBD. 🍷➡️🩺
4/n For example, an ethanol-specific eQTL for LNX2 found only in intestinal epithelial cells is located in an enhancer element and aligns closely with a genetic risk locus for inflammatory bowel disease (IBD),
May 5, 2025 at 7:07 PM
4/n For example, an ethanol-specific eQTL for LNX2 found only in intestinal epithelial cells is located in an enhancer element and aligns closely with a genetic risk locus for inflammatory bowel disease (IBD),
3/n We identified hundreds of response eQTLs, effects change in response to exposure of common environmental factors. These context-dependent eQTLs are often missed in conventional eQTL assays using untreated samples.🌡️🧬
May 5, 2025 at 7:07 PM
3/n We identified hundreds of response eQTLs, effects change in response to exposure of common environmental factors. These context-dependent eQTLs are often missed in conventional eQTL assays using untreated samples.🌡️🧬
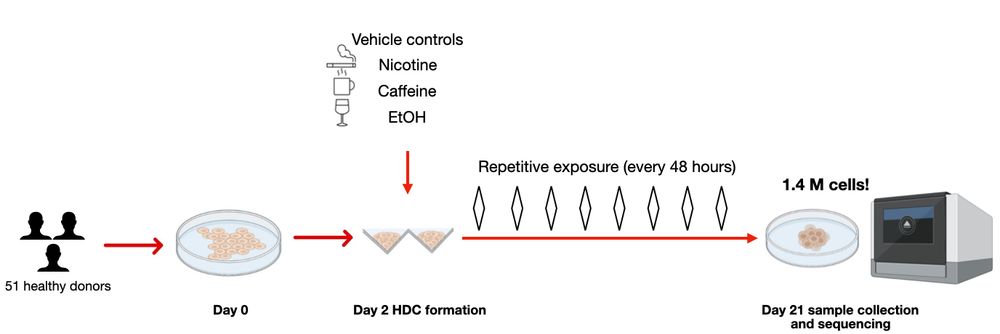
2/n Here, we utilized heterogeneous differentiating cultures (HDCs) to do this. Using single-cell RNA-seq data from 1.4 MILLION cells across 51 genetically diverse individuals, we studied eQTLs under environmental exposures: caffeine, ethanol, and nicotine. ☕️🍷🚬
May 5, 2025 at 7:07 PM
2/n Here, we utilized heterogeneous differentiating cultures (HDCs) to do this. Using single-cell RNA-seq data from 1.4 MILLION cells across 51 genetically diverse individuals, we studied eQTLs under environmental exposures: caffeine, ethanol, and nicotine. ☕️🍷🚬

