
generously supported by the #JeffreyCheahFoundation
Thank you for your support 🙏!!
generously supported by the #JeffreyCheahFoundation
Thank you for your support 🙏!!
And our generous funders 🥹
@cancerresearchuk.org
@nihr.bsky.social
@bcrfcure.bsky.social
#JosefSteiner #GrayFoundation
🙏💕
that enable us to do our research!
/end
And our generous funders 🥹
@cancerresearchuk.org
@nihr.bsky.social
@bcrfcure.bsky.social
#JosefSteiner #GrayFoundation
🙏💕
that enable us to do our research!
/end
This would could not be achieved without the leadership, grit and determination of Gene Koh
Supported by Scott Nanda, Xueqing Zou, Giuseppe Rinaldi
& many more
❤️❤️
12/n
This would could not be achieved without the leadership, grit and determination of Gene Koh
Supported by Scott Nanda, Xueqing Zou, Giuseppe Rinaldi
& many more
❤️❤️
12/n
Consistent in several cohorts.
Using signatures gives much greater specificity for clinical classification.
11/n


Consistent in several cohorts.
Using signatures gives much greater specificity for clinical classification.
11/n
Performance was better than any other classifier or similar ilk.
10/n

Performance was better than any other classifier or similar ilk.
10/n
9/n
9/n
Mechanism proposed therein.
8/n


Mechanism proposed therein.
8/n
10 had features reported by COSMIC and we matched these and kept their nomenclature.
We found 27 new signatures. See paper for details.
7/n

10 had features reported by COSMIC and we matched these and kept their nomenclature.
We found 27 new signatures. See paper for details.
7/n
6/n


6/n
5/n

5/n
all signal is focused on just two channels in COSMIC.
We propose an alternative classification using 5’ and 3’ flanking nucleotides (like subs!), remove cap of 5bp at repeats, amongst others..
resulting in an 89-channel classification system
4/n

all signal is focused on just two channels in COSMIC.
We propose an alternative classification using 5’ and 3’ flanking nucleotides (like subs!), remove cap of 5bp at repeats, amongst others..
resulting in an 89-channel classification system
4/n
They get dumped into COSMIC sigs InD1/InD2 (normal sigs of replication slippage) becoz the COSMIC classification aggregates signal at polynucleotide >5bp into one channel
3/n


They get dumped into COSMIC sigs InD1/InD2 (normal sigs of replication slippage) becoz the COSMIC classification aggregates signal at polynucleotide >5bp into one channel
3/n
ain’t just one entity.
We created isogenic CRISPR KO models of many PRR genes showing clear gene-specific differences (subs & indels)
2/n

ain’t just one entity.
We created isogenic CRISPR KO models of many PRR genes showing clear gene-specific differences (subs & indels)
2/n
academic.oup.com/ced/advance-...
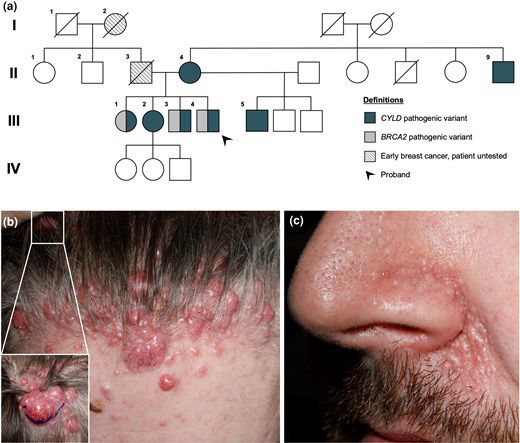
academic.oup.com/ced/advance-...
News right now is unhinged..
News right now is unhinged..

