







We generated new ubiquitous (CAG) and tissue-specific FlpOERT2 lines.
We generated new ubiquitous (CAG) and tissue-specific FlpOERT2 lines.

nature.com/articles/s4159…

nature.com/articles/s4159…
cnic.es/en/convocatori…

cnic.es/en/convocatori…


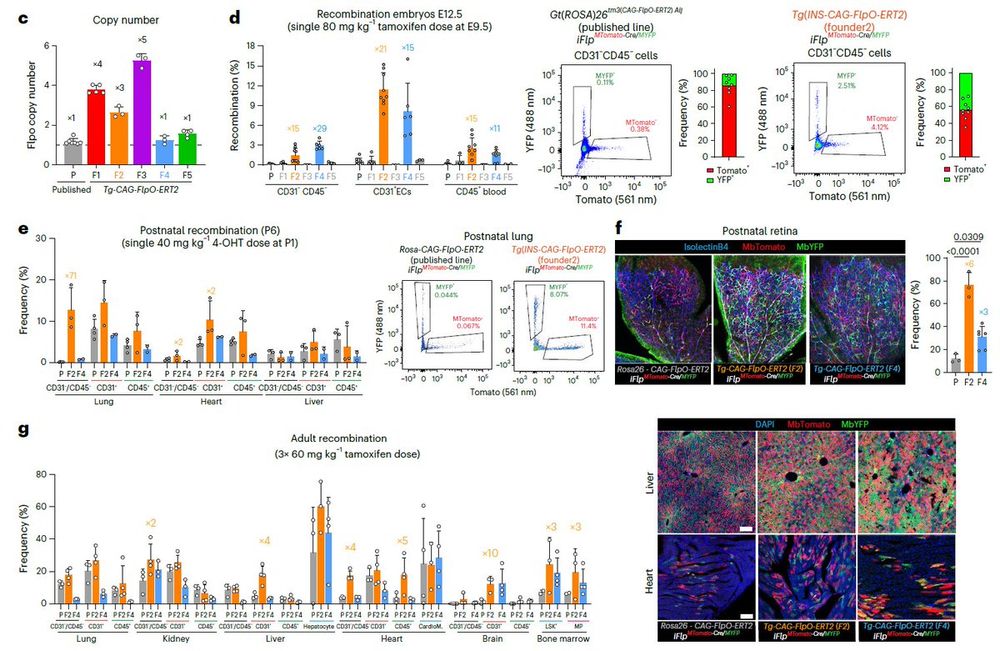
iSuRe-HadCre gave no additional toxicity.
Since 1 dose of 4-OHT is enough to induce iSuRe-HadCre, it is the way fwd for sensitive tissues like below

iSuRe-HadCre gave no additional toxicity.
Since 1 dose of 4-OHT is enough to induce iSuRe-HadCre, it is the way fwd for sensitive tissues like below







x.com/PotenteLab/sta…

x.com/PotenteLab/sta…
rdcu.be/dE5fX

rdcu.be/dE5fX



