You can read our full discussion here: dalmug.org/blood-refere...
You can read our full discussion here: dalmug.org/blood-refere...
The authors also suggest that…
The authors also suggest that…
In this study, the authors characterised the microbiome in blood, saliva and stool of 171 healthy/10 periodontal disease participants.
In this study, the authors characterised the microbiome in blood, saliva and stool of 171 healthy/10 periodontal disease participants.
www.biorxiv.org/content/10.1...
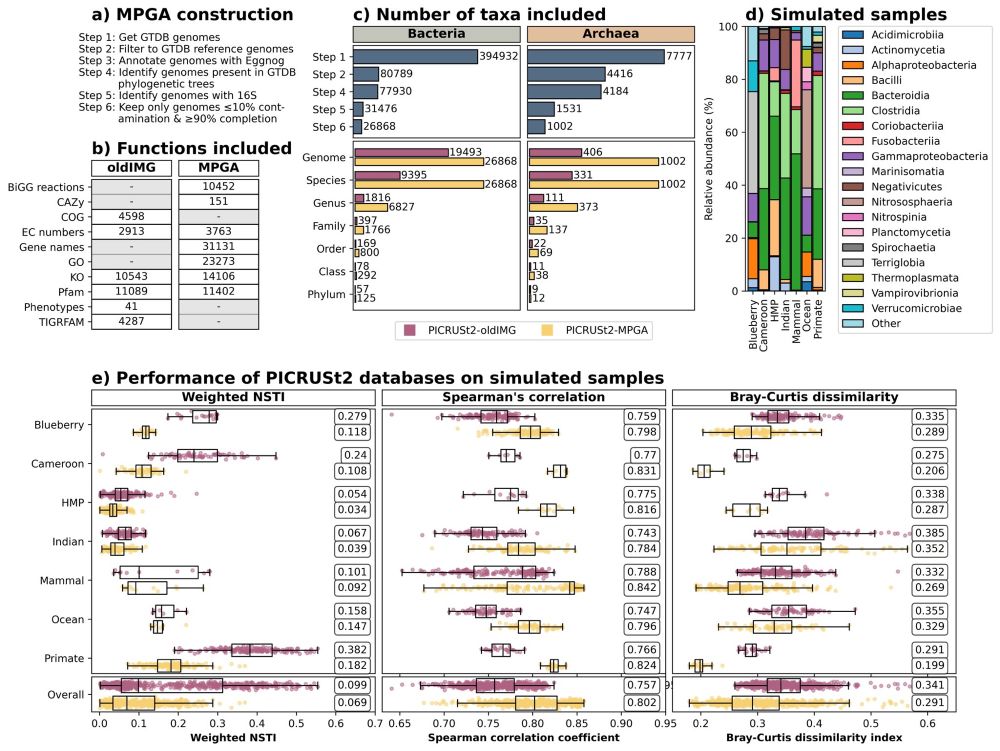
No major changes here: the database used is the same but we’ve added some comparisons of computational resources required and a few extra discussion points.

www.biorxiv.org/content/10.1...
No major changes here: the database used is the same but we’ve added some comparisons of computational resources required and a few extra discussion points.
The new database is included from PICRUSt2 v2.6.0, and if you use it, we’d love to hear your feedback on it!
The new database is included from PICRUSt2 v2.6.0, and if you use it, we’d love to hear your feedback on it!