Head of the https://team.inria.fr/genscale/ at Inria and Irisa.
Algorithmics for sequencing data analyses, genomics and metagenomics.


- Use of minimizers and integer compression for indexing.
- Lower memory footprint and faster query times.
- Minimal impact of false positives on result ranking, using the Rank-Biased Overlap (RBO) metric.
2/8

- Use of minimizers and integer compression for indexing.
- Lower memory footprint and faster query times.
- Minimal impact of false positives on result ranking, using the Rank-Biased Overlap (RBO) metric.
2/8




I'm surprised it has not been described before. Please comment if this is not the case.
Blog post here:
pierrepeterlongo.github.io/2025/03/17/m... 🧪🧬🖥️

I'm surprised it has not been described before. Please comment if this is not the case.
Blog post here:
pierrepeterlongo.github.io/2025/03/17/m... 🧪🧬🖥️
We are hiring
🚨🚨🚨
After the creation of logan-search (see: bsky.app/profile/pier...) we propose a 2-years engineer position for continuing the development and optimizations.
With @rayanchikhi.bsky.social and @tlemane.bsky.social
Details + applications: recrutement.inria.fr/public/class...

We are hiring
🚨🚨🚨
After the creation of logan-search (see: bsky.app/profile/pier...) we propose a 2-years engineer position for continuing the development and optimizations.
With @rayanchikhi.bsky.social and @tlemane.bsky.social
Details + applications: recrutement.inria.fr/public/class...
@yoann.bsky.social
and collaborators.
Reorganize minimizers to allow kmers dichotomic search. That's brilliant.
#bioinformatics 🧬🖥️

@yoann.bsky.social
and collaborators.
Reorganize minimizers to allow kmers dichotomic search. That's brilliant.
#bioinformatics 🧬🖥️
Developed w/ @TeoLemane, & collabs @RayanChikhi, @RNA_Life, Alex Morales, and Luke Pereira

Developed w/ @TeoLemane, & collabs @RayanChikhi, @RNA_Life, Alex Morales, and Luke Pereira
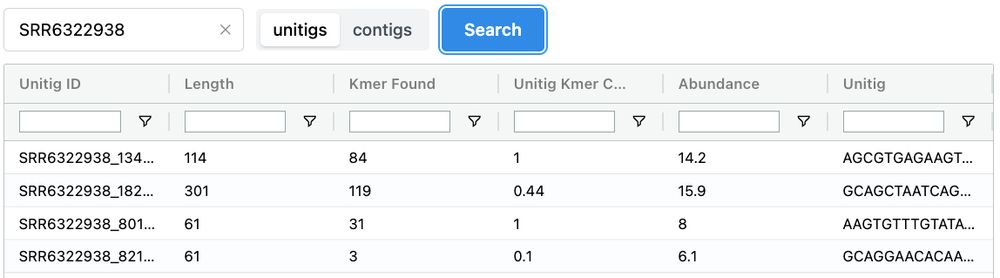
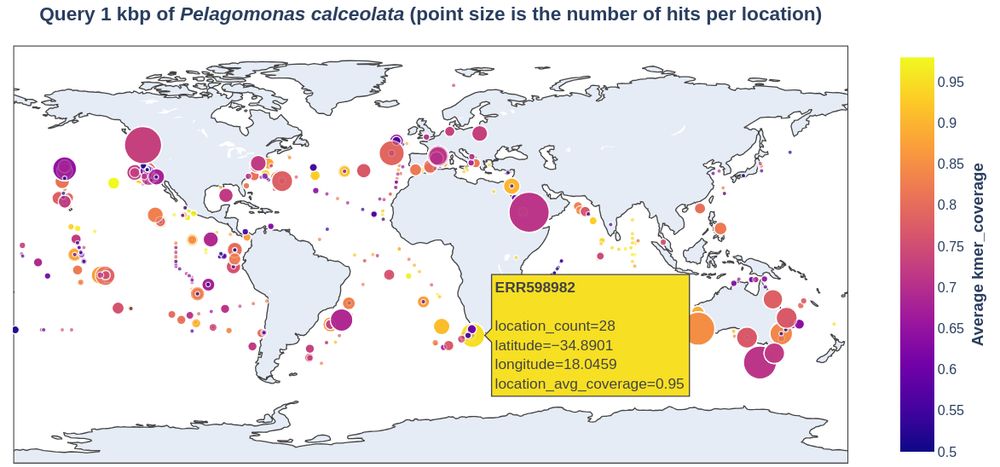

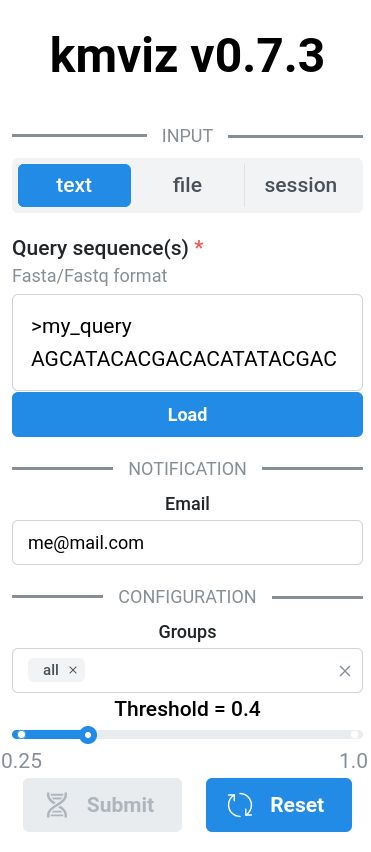

🏔️ logan-search.org 🏔️
#Genomics #Bioinformatics #OpenScience

🏔️ logan-search.org 🏔️
#Genomics #Bioinformatics #OpenScience

