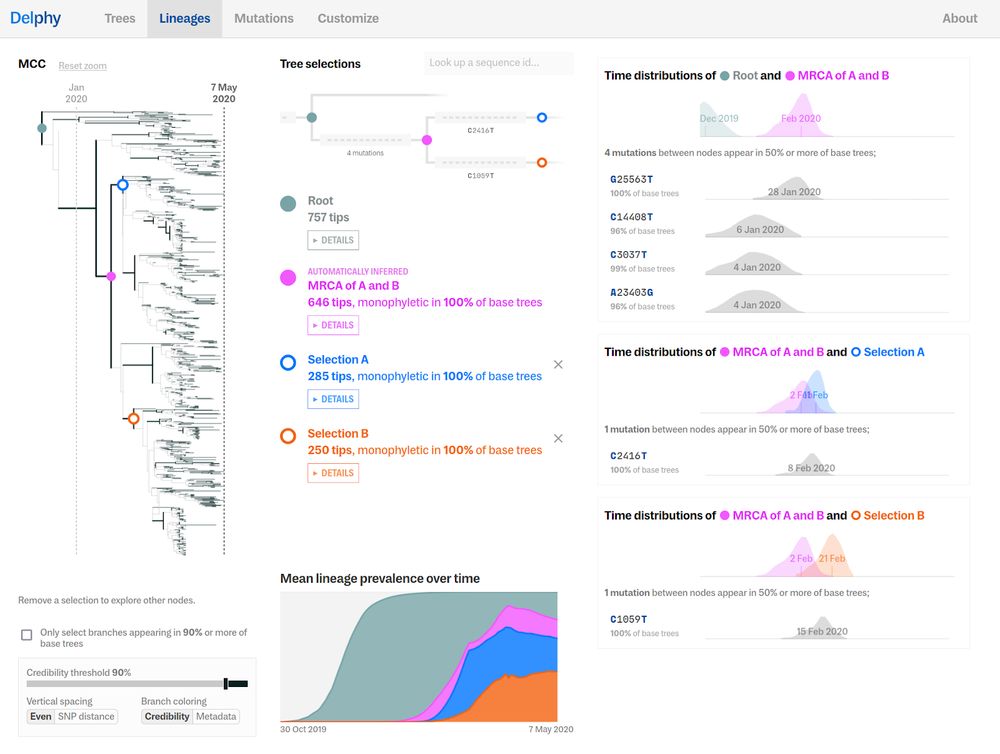
#ScalableTech 12/17

#ScalableTech 12/17
Uses Andersen Lab’s avian flu sequences (1944 as of Mar 22, 2025) and exact dates from GenBank.
Delphy assumes exponential growth—for now.
#OutbreakResponse 11/17

Uses Andersen Lab’s avian flu sequences (1944 as of Mar 22, 2025) and exact dates from GenBank.
Delphy assumes exponential growth—for now.
#OutbreakResponse 11/17
#H5N1 #Phylogenetics #OutbreakResponse 10/17

#H5N1 #Phylogenetics #OutbreakResponse 10/17
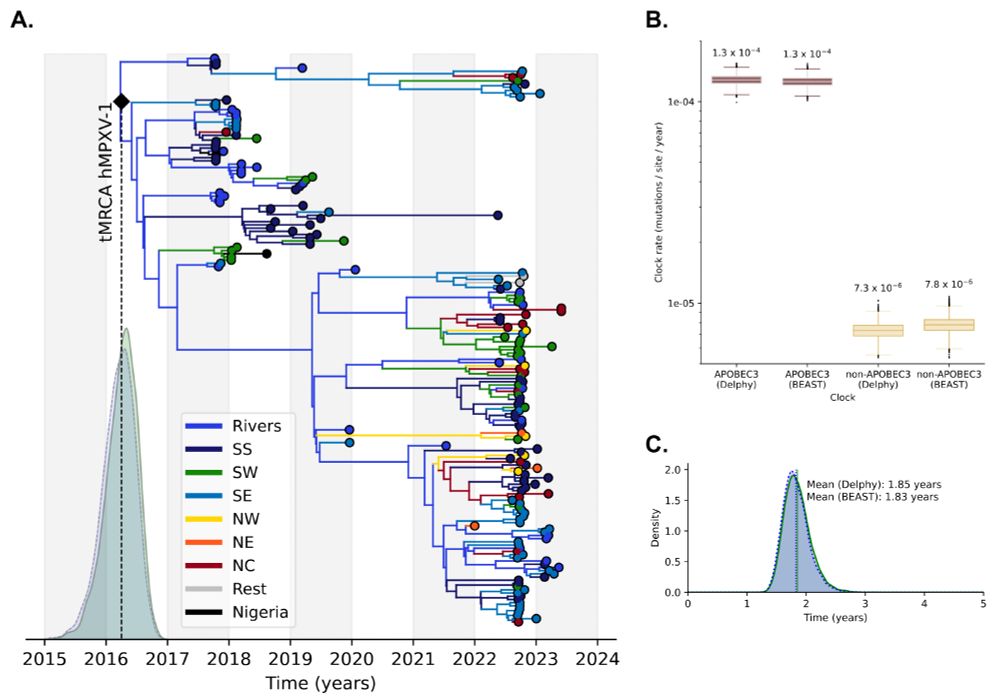
#SARSCoV2 #Phylogenetics #OutbreakResponse 9/17

#SARSCoV2 #Phylogenetics #OutbreakResponse 9/17

In our initial whitepaper, we benchmarked it on viral genome sequence datasets:
🧬 757 SARS-CoV-2
🧬 174 Zika
🧬 81 Ebola
General scaling suggests even greater speedups on larger datasets.
5/17
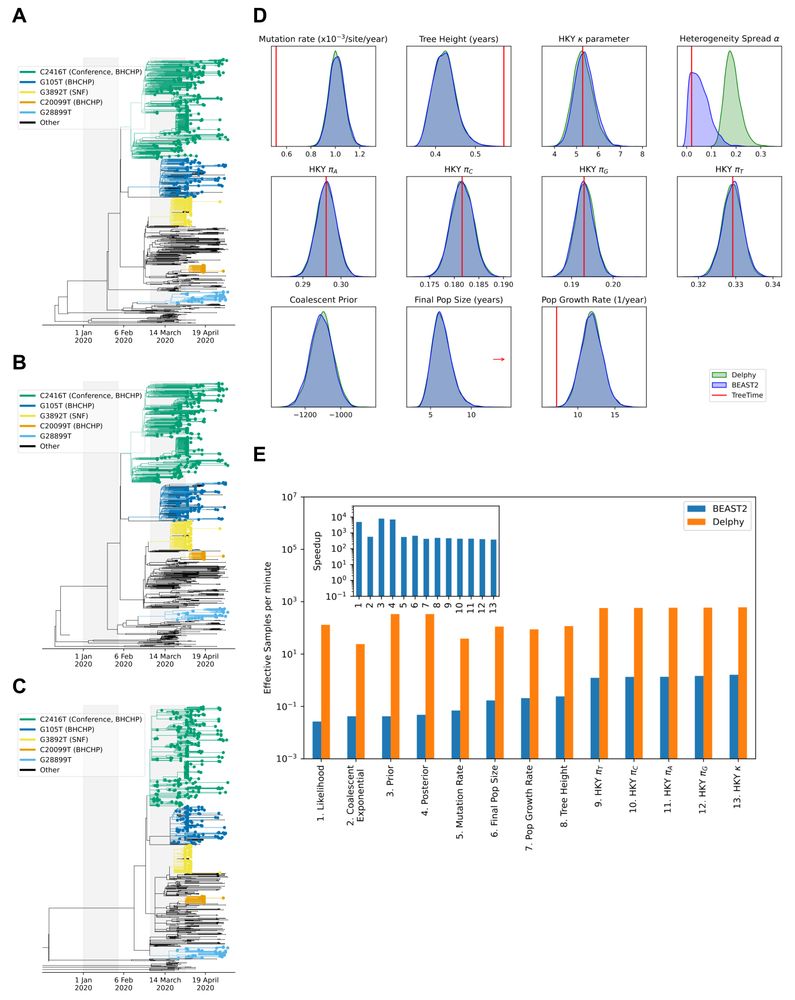
In our initial whitepaper, we benchmarked it on viral genome sequence datasets:
🧬 757 SARS-CoV-2
🧬 174 Zika
🧬 81 Ebola
General scaling suggests even greater speedups on larger datasets.
5/17