▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending
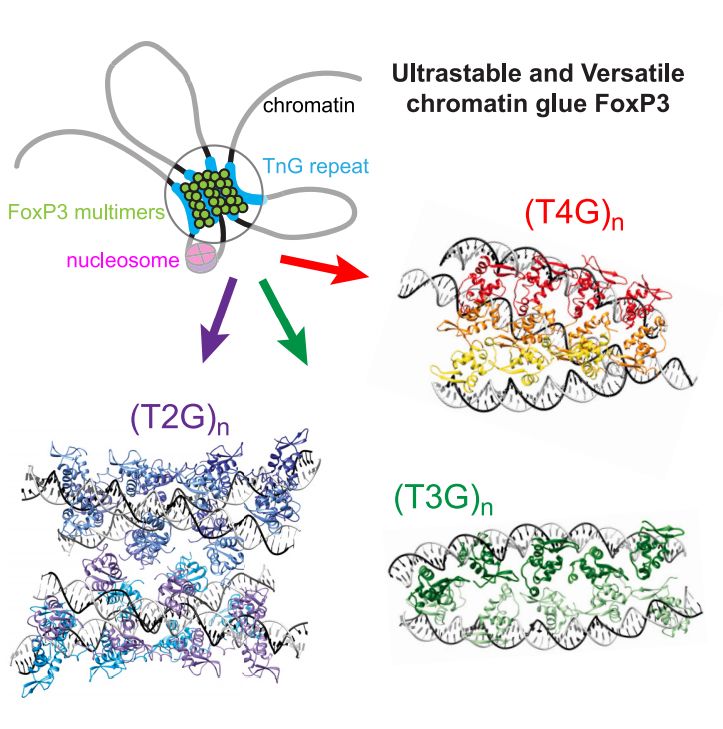
▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending
Really excited to share our latest pre-print on the role of cohesin in homology search, now available on bioRxiv!
www.biorxiv.org/content/10.1...

Really excited to share our latest pre-print on the role of cohesin in homology search, now available on bioRxiv!
www.biorxiv.org/content/10.1...
▶️Measured DNA accessibility in living budding yeast by inducible expression of methyltransferases
▶️The genome is globally accessible in living cells, unlike in isolated nuclei
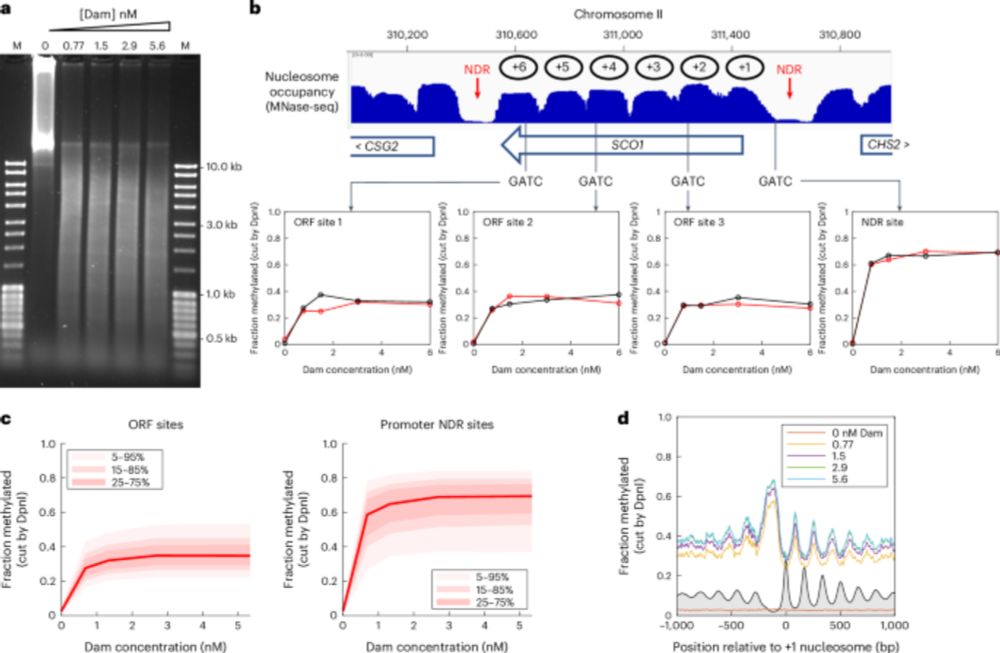
▶️Measured DNA accessibility in living budding yeast by inducible expression of methyltransferases
▶️The genome is globally accessible in living cells, unlike in isolated nuclei

