
Niklas Kempynck
@niklaskemp.bsky.social
PhD Student at the Stein Aerts Lab of Computational Biology. Studying brain genomics
Make sure to also check out the other studies part of the larger effort on identifying and validating enhancer tools.
In the battle against brain disease, researchers can now rely on a new arsenal of genetic tools – The Armamentarium.
Together with scientists from across the NIH BRAIN Initiative, we’ve created and published over 1000 new enhancer AAV vectors.
🧠📈
Together with scientists from across the NIH BRAIN Initiative, we’ve created and published over 1000 new enhancer AAV vectors.
🧠📈
May 21, 2025 at 4:51 PM
Make sure to also check out the other studies part of the larger effort on identifying and validating enhancer tools.
This study was done together with Nelson Johansen and supervised by Trygve Bakken at the @alleninstitute.org. Thanks to all co-authors for the great inter-lab collaboration! Also a personal shoutout to the members in @steinaerts.bsky.social lab for a nice team effort and to Stein for guidance.
May 21, 2025 at 4:48 PM
This study was done together with Nelson Johansen and supervised by Trygve Bakken at the @alleninstitute.org. Thanks to all co-authors for the great inter-lab collaboration! Also a personal shoutout to the members in @steinaerts.bsky.social lab for a nice team effort and to Stein for guidance.
CREsted is available at github.com/aertslab/CRE.... Analysis notebooks can be found at github.com/aertslab/CRE.... All models developed for this preprint and in previous work are available in CREsted through crested.get_model(). We look forward to your feedback!
April 3, 2025 at 2:36 PM
CREsted is available at github.com/aertslab/CRE.... Analysis notebooks can be found at github.com/aertslab/CRE.... All models developed for this preprint and in previous work are available in CREsted through crested.get_model(). We look forward to your feedback!
This was a big collaborative effort, together with @seppedewinter.bsky.social , and with great contributions from @casblaauw.bsky.social , Vasilis and many others. A special shoutout to @lukasmahieu.bsky.social who professionalized the package, and to @steinaerts.bsky.social for supervising.
April 3, 2025 at 2:35 PM
This was a big collaborative effort, together with @seppedewinter.bsky.social , and with great contributions from @casblaauw.bsky.social , Vasilis and many others. A special shoutout to @lukasmahieu.bsky.social who professionalized the package, and to @steinaerts.bsky.social for supervising.
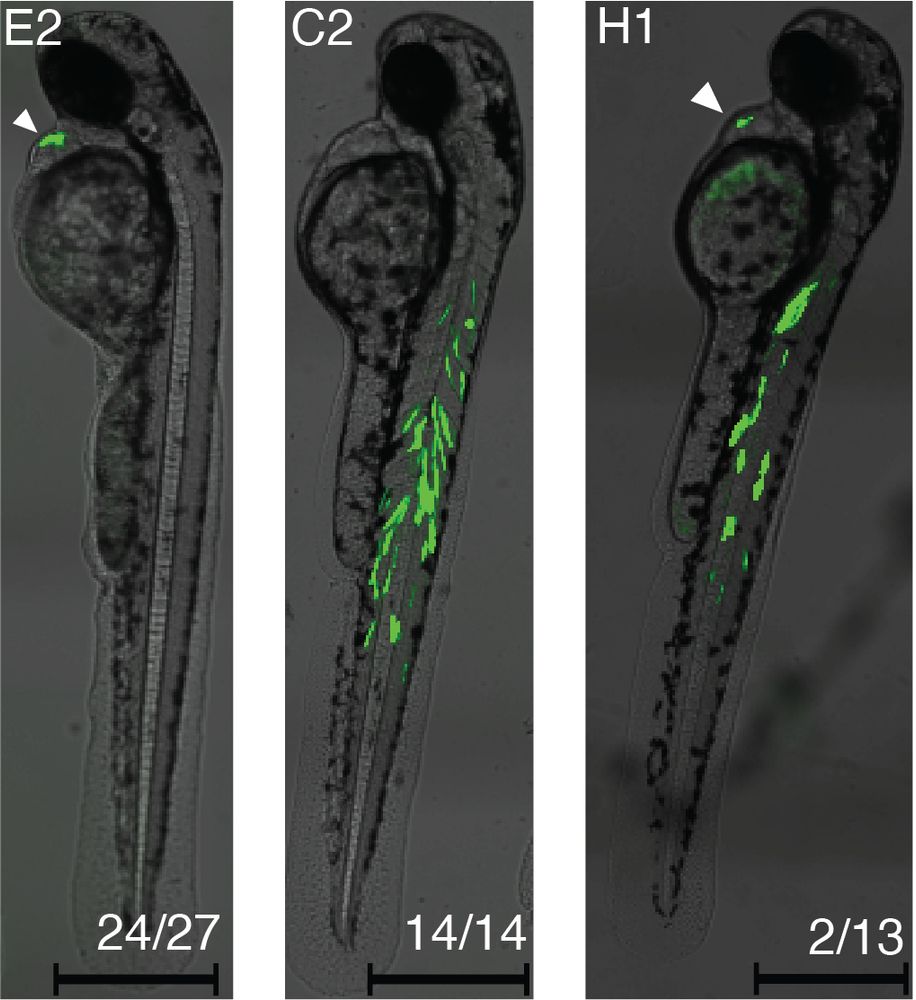
Finally, we train a model on a full-development zebrafish scATAC-seq atlas, and use it to design and in vivo validate cell type- and timepoint-specific enhancers with a high success rate. We also attempt to modulate reporter strength over two cell types.
April 3, 2025 at 2:34 PM
Finally, we train a model on a full-development zebrafish scATAC-seq atlas, and use it to design and in vivo validate cell type- and timepoint-specific enhancers with a high success rate. We also attempt to modulate reporter strength over two cell types.
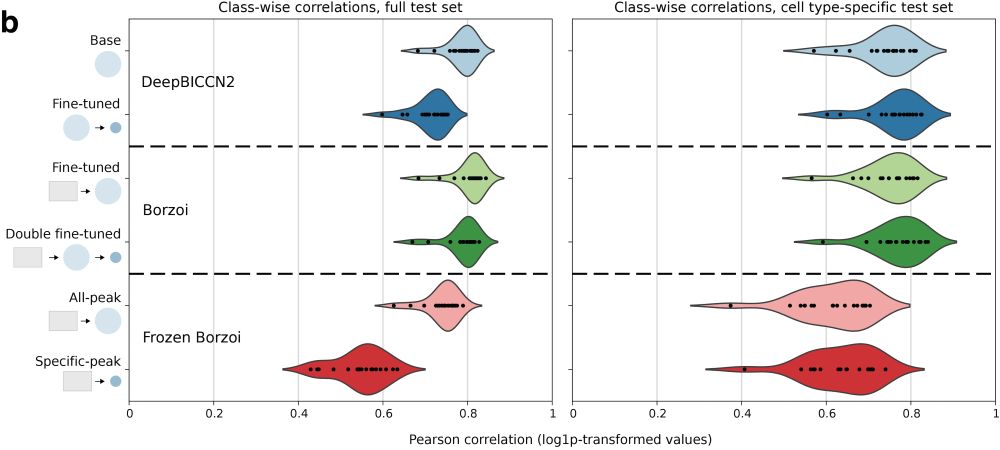
In a new functionality to CREsted, we explore Borzoi fine-tuning to mouse motor cortex scATAC-seq data. We show that fine-tuned models and smaller models from scratch have a near-identical performance.
April 3, 2025 at 2:34 PM
In a new functionality to CREsted, we explore Borzoi fine-tuning to mouse motor cortex scATAC-seq data. We show that fine-tuned models and smaller models from scratch have a near-identical performance.
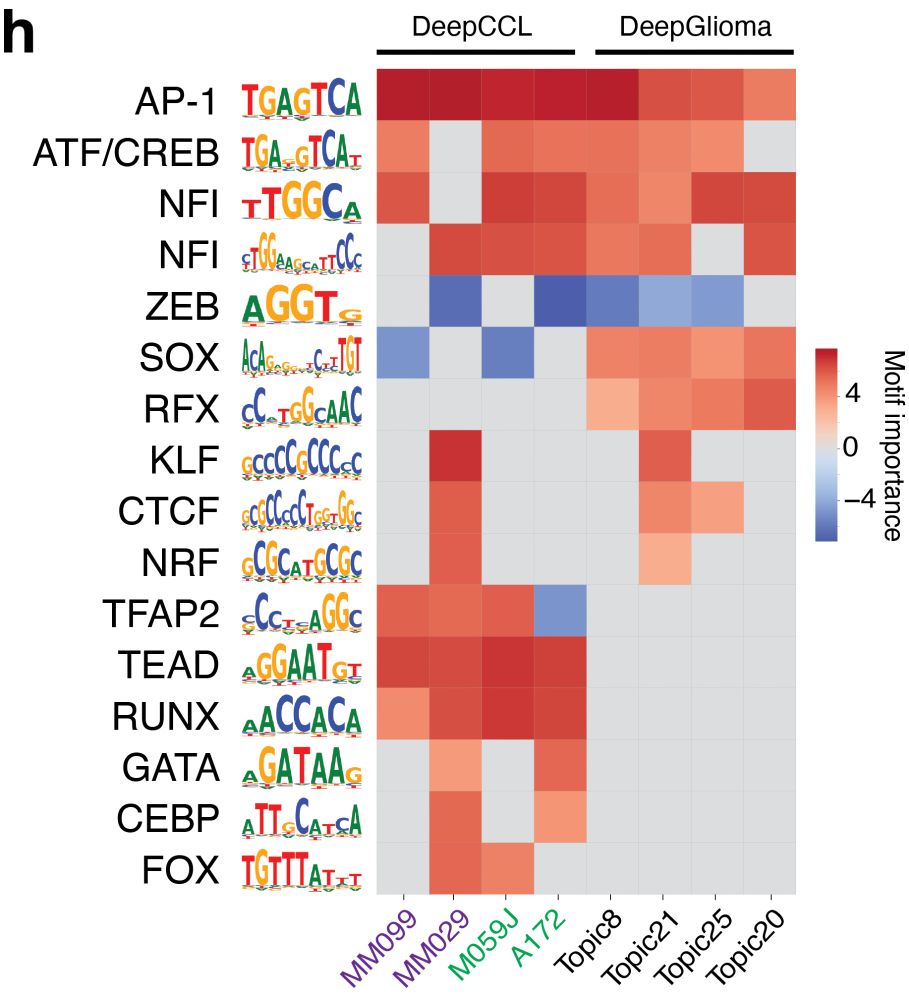
We also study enhancer code inside human cancer cell lines and glioma biopsies and find that enhancer codes between Mesenchymal-like glioblastoma and melanoma states are more similar compared to glioblastoma biopsy data.
April 3, 2025 at 2:33 PM
We also study enhancer code inside human cancer cell lines and glioma biopsies and find that enhancer codes between Mesenchymal-like glioblastoma and melanoma states are more similar compared to glioblastoma biopsy data.
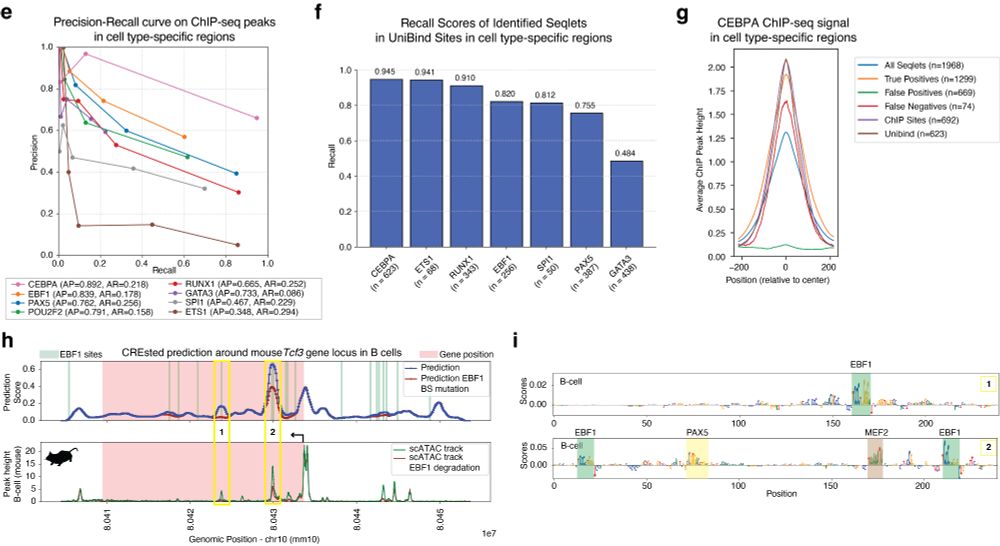
Next, we validated CREsted-identified motif instances from a human PBMC model with ChIP-seq data. We further show that gene locus predictions can be used to simulate the effect of TF degradation on chromatin accessibility.
April 3, 2025 at 2:32 PM
Next, we validated CREsted-identified motif instances from a human PBMC model with ChIP-seq data. We further show that gene locus predictions can be used to simulate the effect of TF degradation on chromatin accessibility.
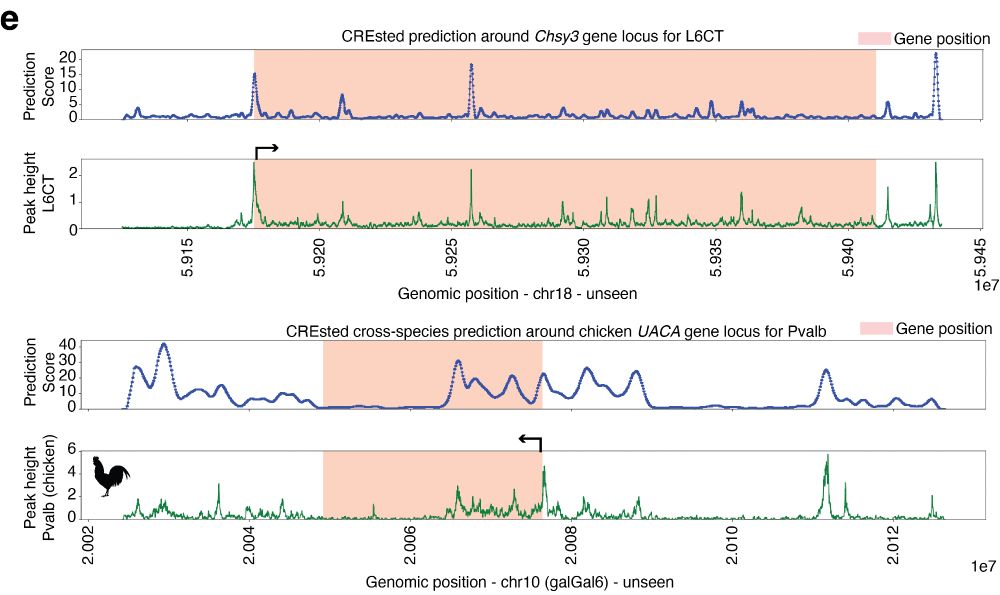
We use the mouse cortex model to highlight CREsted’s gene locus prediction capabilities, both in unseen chromosomes and across species. This presents a powerful tool for potentially annotating genomes across species at high resolution.
April 3, 2025 at 2:32 PM
We use the mouse cortex model to highlight CREsted’s gene locus prediction capabilities, both in unseen chromosomes and across species. This presents a powerful tool for potentially annotating genomes across species at high resolution.
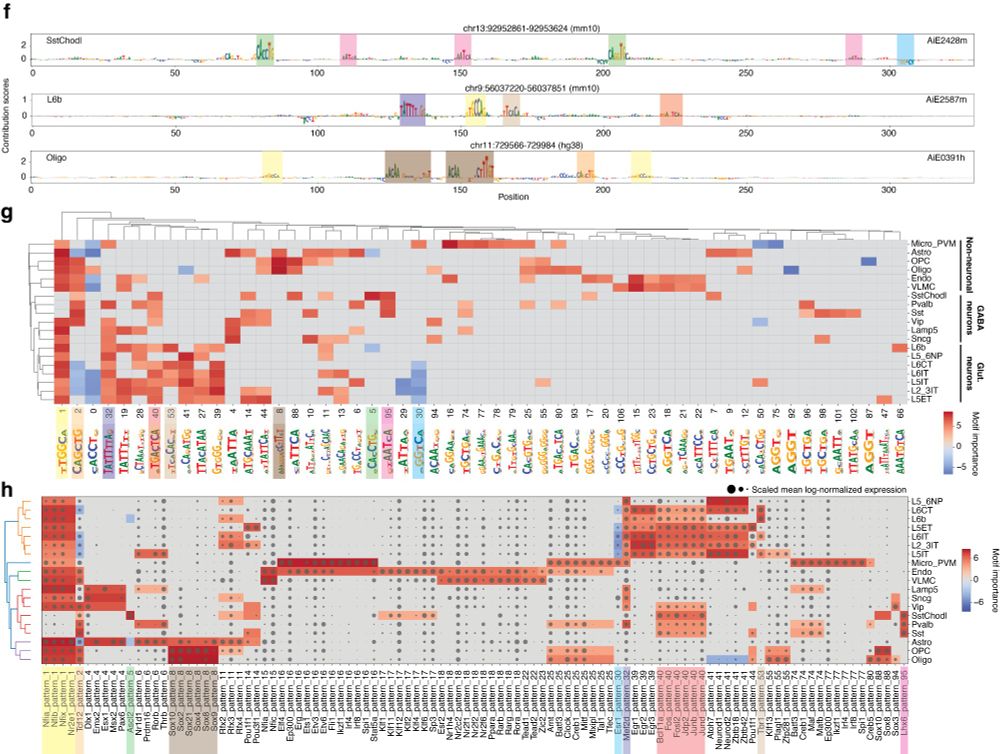
We first demonstrate CREsted’s functionality by providing a complete data-driven analysis of mouse motor cortex enhancer codes across cell types. Through matched scRNA-seq data, we link motifs to likely TF candidates.
April 3, 2025 at 2:31 PM
We first demonstrate CREsted’s functionality by providing a complete data-driven analysis of mouse motor cortex enhancer codes across cell types. Through matched scRNA-seq data, we link motifs to likely TF candidates.
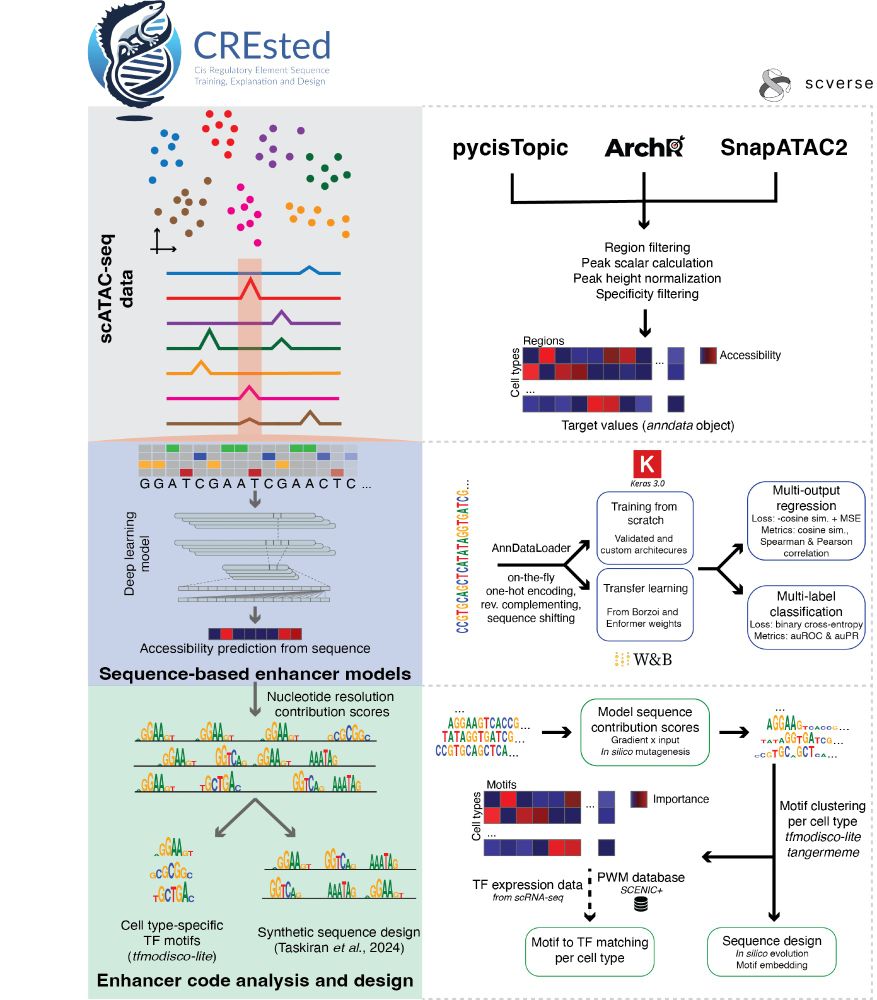
CREsted starts from the outputs of established scATAC preprocessing pipelines, and trains sequence-to-function models on chromatin accessibility per cell type. It provides complete motif analysis tools to infer cell type-specific enhancer codes and holds a comprehensive
enhancer design toolbox.
enhancer design toolbox.
April 3, 2025 at 2:31 PM
CREsted starts from the outputs of established scATAC preprocessing pipelines, and trains sequence-to-function models on chromatin accessibility per cell type. It provides complete motif analysis tools to infer cell type-specific enhancer codes and holds a comprehensive
enhancer design toolbox.
enhancer design toolbox.

