
Read more about our mission at achira.ai
I talked with CEO @jchodera.bsky.social about plans to combine AI- and physics-based methods for drug discovery and moving beyond 45 years of the same type of models behind molecular dynamics.
My latest @endpts.com:
endpts.com/achira-raise...

Read more about our mission at achira.ai
Antibodies seem like ideal candidates for design! 💉
Here’s a quick thread summarising our new review paper on the state of antibody structure prediction. 👇
1/

Antibodies seem like ideal candidates for design! 💉
Here’s a quick thread summarising our new review paper on the state of antibody structure prediction. 👇
1/
polarishub.io/competitions

polarishub.io/competitions
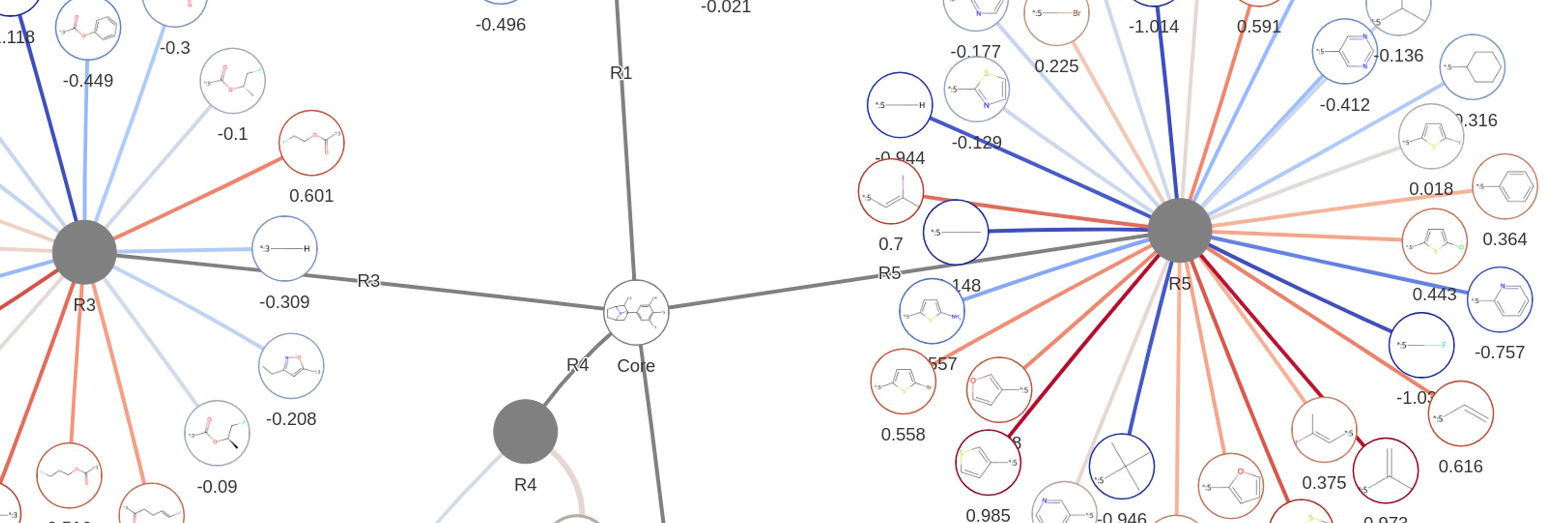
📊 Rapid interpretation of SAR
🔊 Find molecules in the noise
❌ No more lists of molecules
My new favourite visualisation method!
🚀 Read how I visualise Free Wilson analysis here
www.blopig.com/blog/2025/01...

📊 Rapid interpretation of SAR
🔊 Find molecules in the noise
❌ No more lists of molecules
My new favourite visualisation method!
🚀 Read how I visualise Free Wilson analysis here
www.blopig.com/blog/2025/01...

📊 Rapid interpretation of SAR
🔊 Find molecules in the noise
❌ No more lists of molecules
My new favourite visualisation method!
🚀 Read how I visualise Free Wilson analysis here
www.blopig.com/blog/2025/01...
This tool stitches together the atoms of parent hits and re-animates (minimises) them into a conformation close to the parents, preserving their interactions: a must for crystallography-driven drug discovery
doi.org/10.1186/s133...
#chemsky #drugdiscovery

This tool stitches together the atoms of parent hits and re-animates (minimises) them into a conformation close to the parents, preserving their interactions: a must for crystallography-driven drug discovery
doi.org/10.1186/s133...
#chemsky #drugdiscovery

