8/8
8/8
7/8
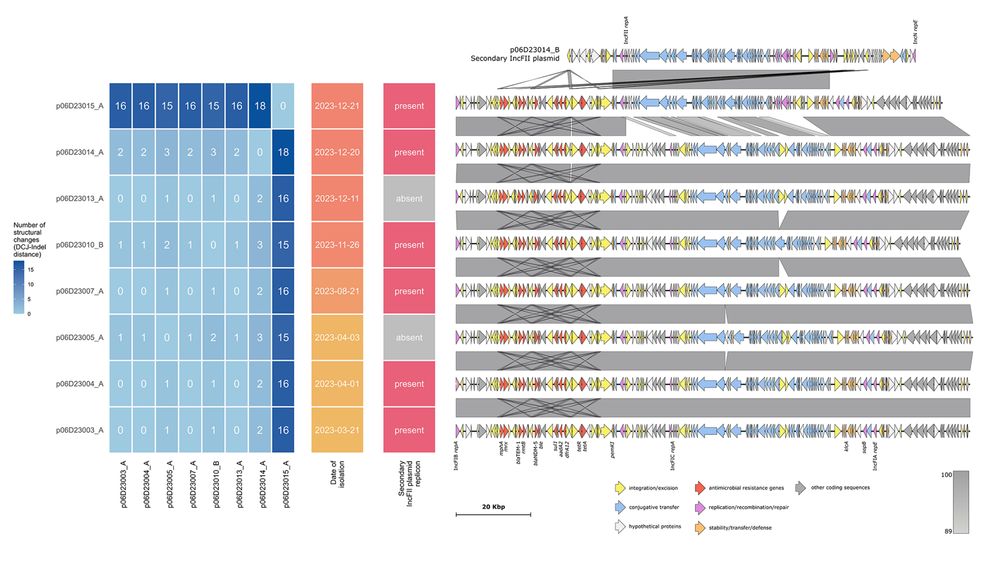
7/8
6/8

6/8
5/8

5/8
4/8
4/8
KPC: journals.asm.org/doi/full/10....
OXA: www.microbiologyresearch.org/content/jour...
3/8

KPC: journals.asm.org/doi/full/10....
OXA: www.microbiologyresearch.org/content/jour...
3/8
2/8
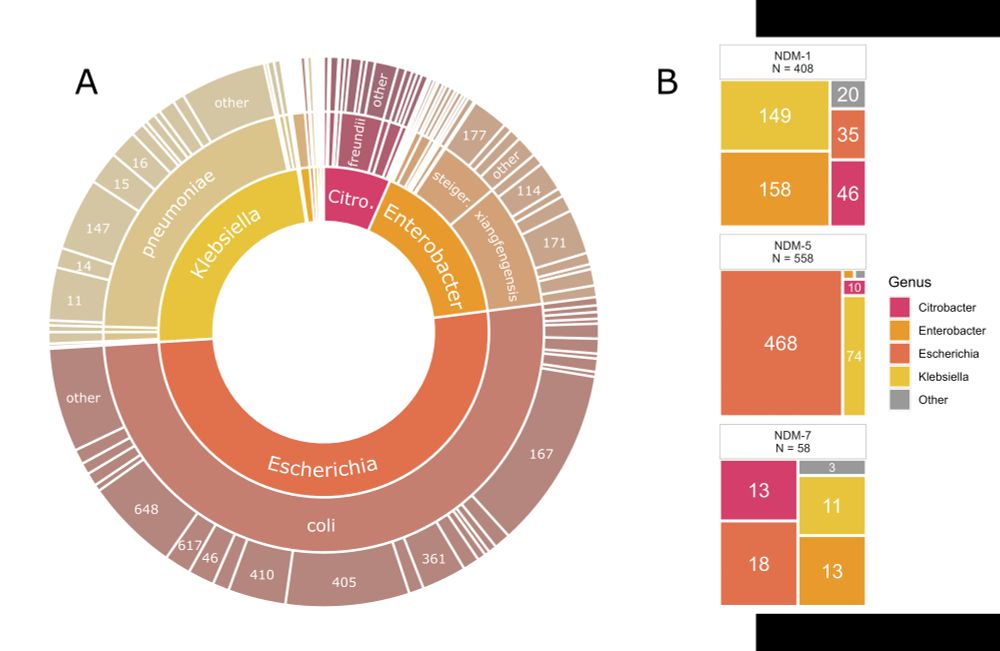
2/8
Feel free to reach out with questions/comments!
3/3
Feel free to reach out with questions/comments!
3/3
2/3
2/3

