Nathaniel Blalock
@nathanielblalock.bsky.social
Graduate Research Assistant in Dr. Philip Romero's Lab at Duke/Wisconsin Reinforcement and Deep Learning for Protein Redesign | He/him
Let me know if you’d like me to clarify anything. I’m happy to talk!
May 25, 2025 at 8:54 PM
Let me know if you’d like me to clarify anything. I’m happy to talk!
Me too 🤪 It is really exciting to be submitting! We definitely learned a lot along the way
May 10, 2025 at 5:27 AM
Me too 🤪 It is really exciting to be submitting! We definitely learned a lot along the way
Thank you for sharing our work @kevinkaichuang.bsky.social! It means a lot
May 10, 2025 at 2:05 AM
Thank you for sharing our work @kevinkaichuang.bsky.social! It means a lot
Thank you for posting about our preprint!
May 8, 2025 at 6:03 PM
Thank you for posting about our preprint!
and our open-source code at github.com/RomeroLab/RLXF

GitHub - RomeroLab/RLXF: Consolidated repository to perform RLXF
Consolidated repository to perform RLXF. Contribute to RomeroLab/RLXF development by creating an account on GitHub.
github.com
May 8, 2025 at 6:02 PM
and our open-source code at github.com/RomeroLab/RLXF
Want to learn more? Check out our preprint at www.biorxiv.org/content/10.1...

Functional alignment of protein language models via reinforcement learning
Protein language models (pLMs) enable generative design of novel protein sequences but remain fundamentally misaligned with protein engineering goals, as they lack explicit understanding of function a...
www.biorxiv.org
May 8, 2025 at 6:02 PM
Want to learn more? Check out our preprint at www.biorxiv.org/content/10.1...
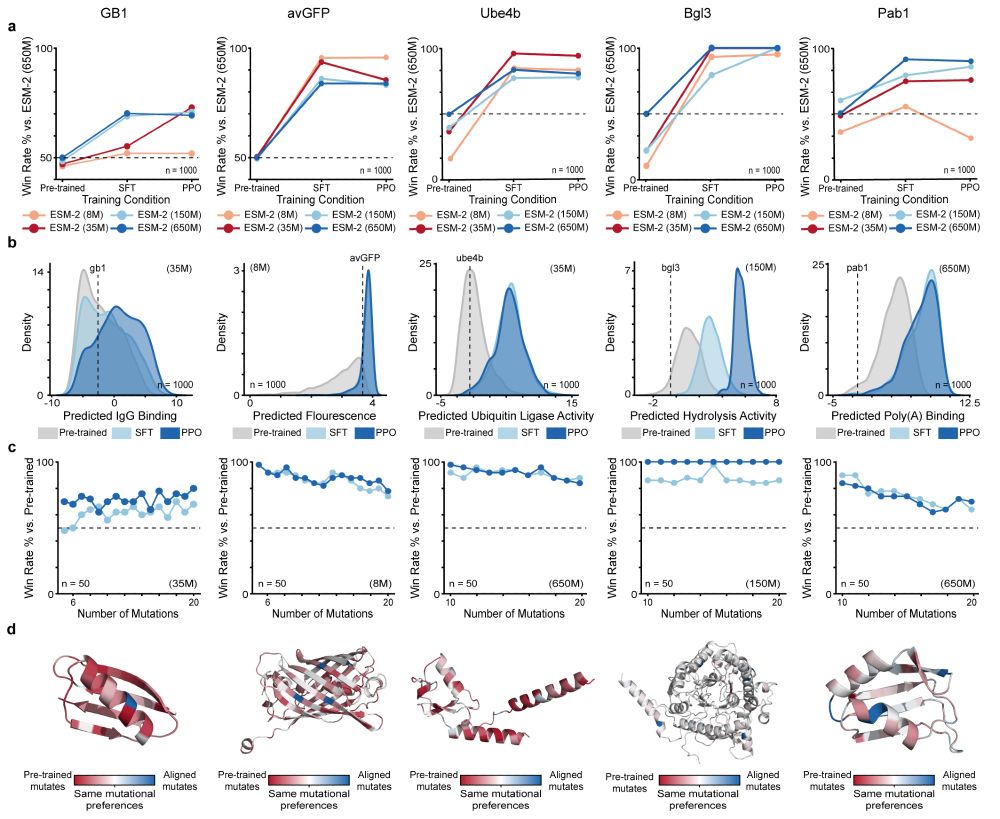
We apply RLXF across five diverse protein classes to demonstrate its generalizability and effectiveness at generating optimized sequences by learning functional constraints beyond those captured during pre-training
May 8, 2025 at 6:02 PM
We apply RLXF across five diverse protein classes to demonstrate its generalizability and effectiveness at generating optimized sequences by learning functional constraints beyond those captured during pre-training
Experimental validation reveals the RLXF-aligned model generates a higher fraction of functional sequences, a greater number of sequences more fluorescent than CreiLOV, and the brightest oxygen-independent fluorescent protein variant reported to date

May 8, 2025 at 6:02 PM
Experimental validation reveals the RLXF-aligned model generates a higher fraction of functional sequences, a greater number of sequences more fluorescent than CreiLOV, and the brightest oxygen-independent fluorescent protein variant reported to date
We align ESM-2 to experimental fluorescence data from the CreiLOV flavin-binding fluorescent protein. The aligned model learns to prioritize mutations that enhance fluorescence, many of which are missed by the base model

May 8, 2025 at 6:02 PM
We align ESM-2 to experimental fluorescence data from the CreiLOV flavin-binding fluorescent protein. The aligned model learns to prioritize mutations that enhance fluorescence, many of which are missed by the base model
RLXF follows a two-phase strategy inspired by RLHF. Supervised Fine-Tuning initializes the model in the right region of sequence space. Proximal Policy Optimization directly aligns sequence generation with feedback from a reward function like a sequence-function predictor

May 8, 2025 at 6:02 PM
RLXF follows a two-phase strategy inspired by RLHF. Supervised Fine-Tuning initializes the model in the right region of sequence space. Proximal Policy Optimization directly aligns sequence generation with feedback from a reward function like a sequence-function predictor
Pre-trained pLMs generate highly diverse sequences mirroring statistical patterns from natural proteins. But here's the challenge: they lack an explicit understanding of function, often failing to generate proteins with enhanced or non-natural activities. RLXF bridges this gap!
May 8, 2025 at 6:02 PM
Pre-trained pLMs generate highly diverse sequences mirroring statistical patterns from natural proteins. But here's the challenge: they lack an explicit understanding of function, often failing to generate proteins with enhanced or non-natural activities. RLXF bridges this gap!
It was a pleasure meeting you! Y'all are doing super interesting and relevant work. It will be cool to see how we can continue to interact and maybe collaborate in the future!
December 20, 2024 at 8:50 PM
It was a pleasure meeting you! Y'all are doing super interesting and relevant work. It will be cool to see how we can continue to interact and maybe collaborate in the future!
Favorite foods! Tandoori chicken and chili momo's: everestkitchen.ca. Onigiri! www.onigiriya.ca. Pho: www.viethouserestaurant.com.



December 20, 2024 at 4:59 PM
Favorite foods! Tandoori chicken and chili momo's: everestkitchen.ca. Onigiri! www.onigiriya.ca. Pho: www.viethouserestaurant.com.
Papers #4: arxiv.org/abs/2406.17692 from the incredible
@gregdnlp.bsky.social. I really like how explore what happens during the alignment of LLM's with RLHF. This was so cool to see having observed similar outcomes in my research.
@gregdnlp.bsky.social. I really like how explore what happens during the alignment of LLM's with RLHF. This was so cool to see having observed similar outcomes in my research.

From Distributional to Overton Pluralism: Investigating Large Language Model Alignment
The alignment process changes several properties of a large language model's (LLM's) output distribution. We analyze two aspects of post-alignment distributional shift of LLM responses. First, we re-e...
arxiv.org
December 20, 2024 at 4:54 PM
Papers #4: arxiv.org/abs/2406.17692 from the incredible
@gregdnlp.bsky.social. I really like how explore what happens during the alignment of LLM's with RLHF. This was so cool to see having observed similar outcomes in my research.
@gregdnlp.bsky.social. I really like how explore what happens during the alignment of LLM's with RLHF. This was so cool to see having observed similar outcomes in my research.
Papers #2-3: arxiv.org/abs/2402.10210 and arxiv.org/abs/2405.00675 from the incredible
@quanquangu.bsky.social. I really like how they explore new techniques for RLHF
@quanquangu.bsky.social. I really like how they explore new techniques for RLHF

Self-Play Fine-Tuning of Diffusion Models for Text-to-Image Generation
Fine-tuning Diffusion Models remains an underexplored frontier in generative artificial intelligence (GenAI), especially when compared with the remarkable progress made in fine-tuning Large Language M...
arxiv.org
December 20, 2024 at 4:53 PM
Papers #2-3: arxiv.org/abs/2402.10210 and arxiv.org/abs/2405.00675 from the incredible
@quanquangu.bsky.social. I really like how they explore new techniques for RLHF
@quanquangu.bsky.social. I really like how they explore new techniques for RLHF
Paper #1: arxiv.org/abs/2412.12979
Aligning autoregressive pLM's to generate EGFR binders via Direct Policy Optimization (DPO) from the incredible @noeliaferruz.bsky.social who gave a great talk as part of the MLSB workshop
Aligning autoregressive pLM's to generate EGFR binders via Direct Policy Optimization (DPO) from the incredible @noeliaferruz.bsky.social who gave a great talk as part of the MLSB workshop

Guiding Generative Protein Language Models with Reinforcement Learning
Autoregressive protein language models (pLMs) have emerged as powerful tools to efficiently design functional proteins with extraordinary diversity, as evidenced by the successful generation of divers...
arxiv.org
December 20, 2024 at 4:42 PM
Paper #1: arxiv.org/abs/2412.12979
Aligning autoregressive pLM's to generate EGFR binders via Direct Policy Optimization (DPO) from the incredible @noeliaferruz.bsky.social who gave a great talk as part of the MLSB workshop
Aligning autoregressive pLM's to generate EGFR binders via Direct Policy Optimization (DPO) from the incredible @noeliaferruz.bsky.social who gave a great talk as part of the MLSB workshop
Hey Kevin, could I be added? This is really helpful for joining Bluesky! Thank you for doing it
December 17, 2024 at 6:38 PM
Hey Kevin, could I be added? This is really helpful for joining Bluesky! Thank you for doing it

