

@mukeshbhendarkar.bsky.social
@mukeshbhendarkar.bsky.social





"Global distribution patterns of siphonophores across horizontal and vertical oceanic gradients" by reanalyzing data from the Malaspina expedition.

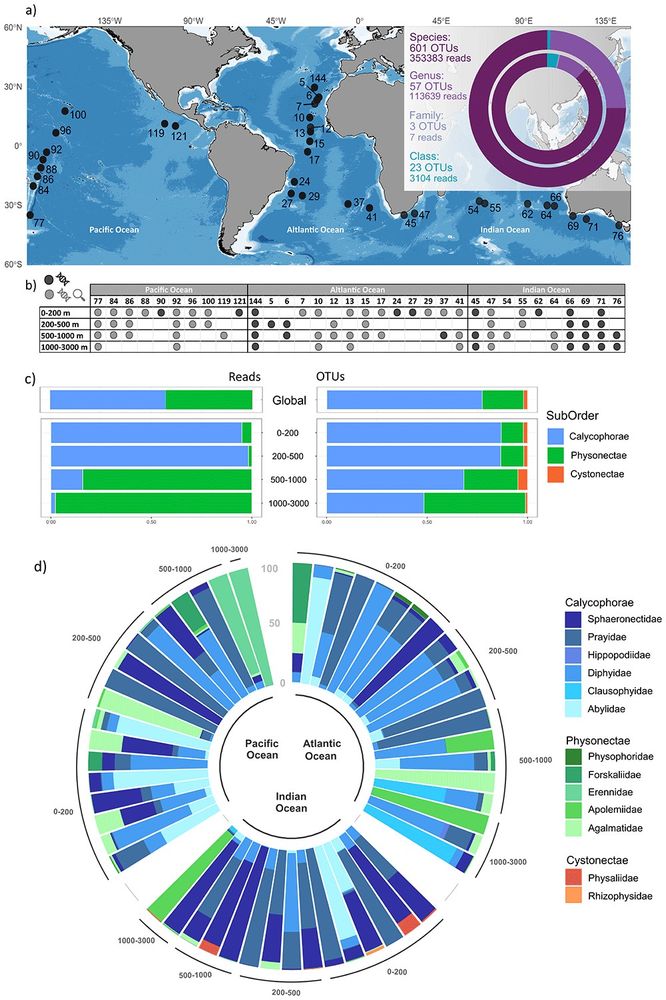
"Global distribution patterns of siphonophores across horizontal and vertical oceanic gradients" by reanalyzing data from the Malaspina expedition.
Registration for @TheFSBI symposium is now open!!
Key dates:
Symposium Award application deadline: 1st April
Early bird and presenter registration closes: 1st May
Late registration closes: 17th June
fsbi.org.uk/symposium-20...
#FSBI2024

Registration for @TheFSBI symposium is now open!!
Key dates:
Symposium Award application deadline: 1st April
Early bird and presenter registration closes: 1st May
Late registration closes: 17th June
fsbi.org.uk/symposium-20...
#FSBI2024






FSBI Symposium is now OPEN!
Join us for a fantastic gathering in #Bilbao hosted by @azti.bsky.social and featuring leading names in fisheries genomics
fsbi.org.uk/symposium-20...

FSBI Symposium is now OPEN!
Join us for a fantastic gathering in #Bilbao hosted by @azti.bsky.social and featuring leading names in fisheries genomics
fsbi.org.uk/symposium-20...








Sophie von der Heyden from Stellenbosch University in South Africa utilizes #genetic and #genomic approaches for studying various aspects of marine and freshwater systems
Don't miss it!
bit.ly/3rV6fft

Sophie von der Heyden from Stellenbosch University in South Africa utilizes #genetic and #genomic approaches for studying various aspects of marine and freshwater systems
Don't miss it!
bit.ly/3rV6fft


Read more at ⏩ bit.ly/3rV6fft

Read more at ⏩ bit.ly/3rV6fft
👩🔬 Suzana Musilova (Charles University of Prague) works on the #evolution of fish sensory systems, focusing on different groups of fish from the deep-sea lineages.
Learn more at ▶bit.ly/3rV6fft

👩🔬 Suzana Musilova (Charles University of Prague) works on the #evolution of fish sensory systems, focusing on different groups of fish from the deep-sea lineages.
Learn more at ▶bit.ly/3rV6fft

