Imperial College London, Reader
Website: monotockylab.github.io
GitHub: github.com/MonoTockyLab

✅ConvNet + Grad-CAM decodes fluorescent timer (Tocky) data
✅Enables single-cell identification of enhancer-dependent Foxp3 transcription dynamics
www.nature.com/articles/s41...
🔥A step toward data-driven immunology!
www.telegraph.co.uk/science/canc...

www.telegraph.co.uk/science/canc...
www.telegraph.co.uk/science/canc...

www.telegraph.co.uk/science/canc...
www.nature.com/articles/s41...
👉 We build on our original Tocky method by integrating CRISPR & deep learning to decode transcriptional dynamics.
🔥 7 years of work - toward data-driven immunology!

www.nature.com/articles/s41...
👉 We build on our original Tocky method by integrating CRISPR & deep learning to decode transcriptional dynamics.
🔥 7 years of work - toward data-driven immunology!
🔗 onlinelibrary.wiley.com/doi/10.1002/...
GatingTree enables systematic analysis of high-dimensional flow cytometry data, without relying on dimensionality reduction. 💻

🔗 onlinelibrary.wiley.com/doi/10.1002/...
GatingTree enables systematic analysis of high-dimensional flow cytometry data, without relying on dimensionality reduction. 💻
Our ConvNet + Grad-CAM approach decodes Fluorescent Timer (Tocky) data at single-cell resolution.
Using Foxp3 Tocky, we reveal CNS2-controlled and developmental transcriptional dynamics.
Built on 7 years of work with AI and CRISPR!
www.nature.com/articles/s41...

Our ConvNet + Grad-CAM approach decodes Fluorescent Timer (Tocky) data at single-cell resolution.
Using Foxp3 Tocky, we reveal CNS2-controlled and developmental transcriptional dynamics.
Built on 7 years of work with AI and CRISPR!
www.nature.com/articles/s41...
GatingTree skips such methods, directly analyses high-dimensional data, builds reproducible tree paths across markers, and reveals group-specific effects.
No dimensionality reduction, just interpretable gates. 🌳

GatingTree skips such methods, directly analyses high-dimensional data, builds reproducible tree paths across markers, and reveals group-specific effects.
No dimensionality reduction, just interpretable gates. 🌳
🔗 onlinelibrary.wiley.com/doi/10.1002/...
GatingTree enables systematic analysis of high-dimensional flow cytometry data, without relying on dimensionality reduction. 💻

🔗 onlinelibrary.wiley.com/doi/10.1002/...
GatingTree enables systematic analysis of high-dimensional flow cytometry data, without relying on dimensionality reduction. 💻
✅ConvNet + Grad-CAM decodes fluorescent timer (Tocky) data
✅Enables single-cell identification of enhancer-dependent Foxp3 transcription dynamics
www.nature.com/articles/s41...
🔥A step toward data-driven immunology!

✅ConvNet + Grad-CAM decodes fluorescent timer (Tocky) data
✅Enables single-cell identification of enhancer-dependent Foxp3 transcription dynamics
www.nature.com/articles/s41...
🔥A step toward data-driven immunology!
✅ConvNet + Grad-CAM decodes fluorescent timer (Tocky) data
✅Enables single-cell identification of enhancer-dependent Foxp3 transcription dynamics
www.nature.com/articles/s41...
🔥A step toward data-driven immunology!

✅ConvNet + Grad-CAM decodes fluorescent timer (Tocky) data
✅Enables single-cell identification of enhancer-dependent Foxp3 transcription dynamics
www.nature.com/articles/s41...
🔥A step toward data-driven immunology!
Our ConvNet + Grad-CAM approach decodes Fluorescent Timer (Tocky) data at single-cell resolution.
Using Foxp3 Tocky, we reveal CNS2-controlled and developmental transcriptional dynamics.
Built on 7 years of work with AI and CRISPR!
www.nature.com/articles/s41...

Our ConvNet + Grad-CAM approach decodes Fluorescent Timer (Tocky) data at single-cell resolution.
Using Foxp3 Tocky, we reveal CNS2-controlled and developmental transcriptional dynamics.
Built on 7 years of work with AI and CRISPR!
www.nature.com/articles/s41...
The start of our collaboration goes back to my own Runx paper in 2007: www.nature.com/articles/nat...
This Runx/HTLV-1 study is the latest milestone 🔬.
Honoured to be a co-author 📄✨
An intragenic open chromatin region in human T cell leukemia virus type 1 functions as a transcriptional silencer and regulates transcriptional burst by recruiting the host transcription factor RUNX.
#MicroSky 🧬🦠
www.nature.com/articles/s41...

The start of our collaboration goes back to my own Runx paper in 2007: www.nature.com/articles/nat...
This Runx/HTLV-1 study is the latest milestone 🔬.
Honoured to be a co-author 📄✨
The start of our collaboration goes back to my own Runx paper in 2007: www.nature.com/articles/nat...
This Runx/HTLV-1 study is the latest milestone 🔬.
Honoured to be a co-author 📄✨
An intragenic open chromatin region in human T cell leukemia virus type 1 functions as a transcriptional silencer and regulates transcriptional burst by recruiting the host transcription factor RUNX.
#MicroSky 🧬🦠
www.nature.com/articles/s41...

The start of our collaboration goes back to my own Runx paper in 2007: www.nature.com/articles/nat...
This Runx/HTLV-1 study is the latest milestone 🔬.
Honoured to be a co-author 📄✨
In this work, we integrate this idea with our recent findings: Adult T cell leukemia (ATL) is not 'Treg cancer', challenging the current concept of Treg as a distinct entity.🧵(1/4)
academic.oup.com/discovimmuno...
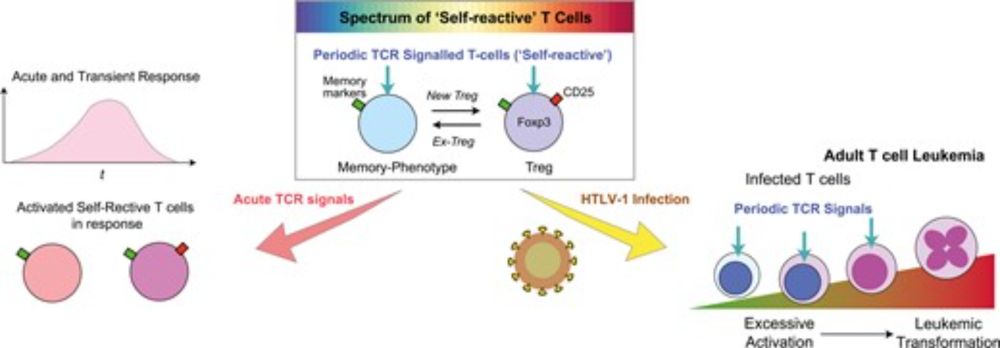
In this work, we integrate this idea with our recent findings: Adult T cell leukemia (ATL) is not 'Treg cancer', challenging the current concept of Treg as a distinct entity.🧵(1/4)
academic.oup.com/discovimmuno...
In this work, we integrate this idea with our recent findings: Adult T cell leukemia (ATL) is not 'Treg cancer', challenging the current concept of Treg as a distinct entity.🧵(1/4)
academic.oup.com/discovimmuno...

In this work, we integrate this idea with our recent findings: Adult T cell leukemia (ATL) is not 'Treg cancer', challenging the current concept of Treg as a distinct entity.🧵(1/4)
academic.oup.com/discovimmuno...
📄 Ono & Satou (2024)
🧬 bit.ly/42gRcuR

📄 Ono & Satou (2024)
🧬 bit.ly/42gRcuR
monotockylab.github.io/blog/2025/As...
The blog explores what is certain in Treg immunology and what is not, unraveling the past two decades of my research life that have been triggered by the non-reproducible evidence in Treg biology.

monotockylab.github.io/blog/2025/As...
The blog explores what is certain in Treg immunology and what is not, unraveling the past two decades of my research life that have been triggered by the non-reproducible evidence in Treg biology.
The study fills key gaps in Treg biology and viral #immunology.
🔗 bit.ly/42gRcuR

The study fills key gaps in Treg biology and viral #immunology.
🔗 bit.ly/42gRcuR
monotockylab.github.io/blog/2025/As...
The blog explores what is certain in Treg immunology and what is not, unraveling the past two decades of my research life that have been triggered by the non-reproducible evidence in Treg biology.

monotockylab.github.io/blog/2025/As...
The blog explores what is certain in Treg immunology and what is not, unraveling the past two decades of my research life that have been triggered by the non-reproducible evidence in Treg biology.
The TockyPrep R package automates and standardizes flow cytometric analysis of Fluorescent Timer reporters, unlocking the analysis of Nr4a3 Tocky mice and other Timer reporters🐭

The TockyPrep R package automates and standardizes flow cytometric analysis of Fluorescent Timer reporters, unlocking the analysis of Nr4a3 Tocky mice and other Timer reporters🐭
The TockyPrep R package automates and standardizes flow cytometric analysis of Fluorescent Timer reporters, unlocking the analysis of Nr4a3 Tocky mice and other Timer reporters🐭

The TockyPrep R package automates and standardizes flow cytometric analysis of Fluorescent Timer reporters, unlocking the analysis of Nr4a3 Tocky mice and other Timer reporters🐭
monotockylab.github.io/TockyPrep/in...
Now featuring an Interactive application to fine-tune your analysis! 🛠️
monotockylab.github.io/TockyPrep/ar...

monotockylab.github.io/TockyPrep/in...
Now featuring an Interactive application to fine-tune your analysis! 🛠️
monotockylab.github.io/TockyPrep/ar...
Read this Techniques and Resources Article by Masahiro Ono @monotockylab.bsky.social and Tessa Crompton:
https://doi.org/10.1242/dev.204255

Read this Techniques and Resources Article by Masahiro Ono @monotockylab.bsky.social and Tessa Crompton:
https://doi.org/10.1242/dev.204255
#Books
#BookSky💙📚
#BookChallenge
Day 20

#Books
#BookSky💙📚
#BookChallenge
Day 20


