Moffitt Lab
@moffittlab.bsky.social
Academic research lab passionate about building the next generation of genomic microscopy methods to reveal unknown features of the interface between microbe and host |
@bostonchildrens.bsky.social & @harvardmed.bsky.social | moffittlab.github.io
@bostonchildrens.bsky.social & @harvardmed.bsky.social | moffittlab.github.io
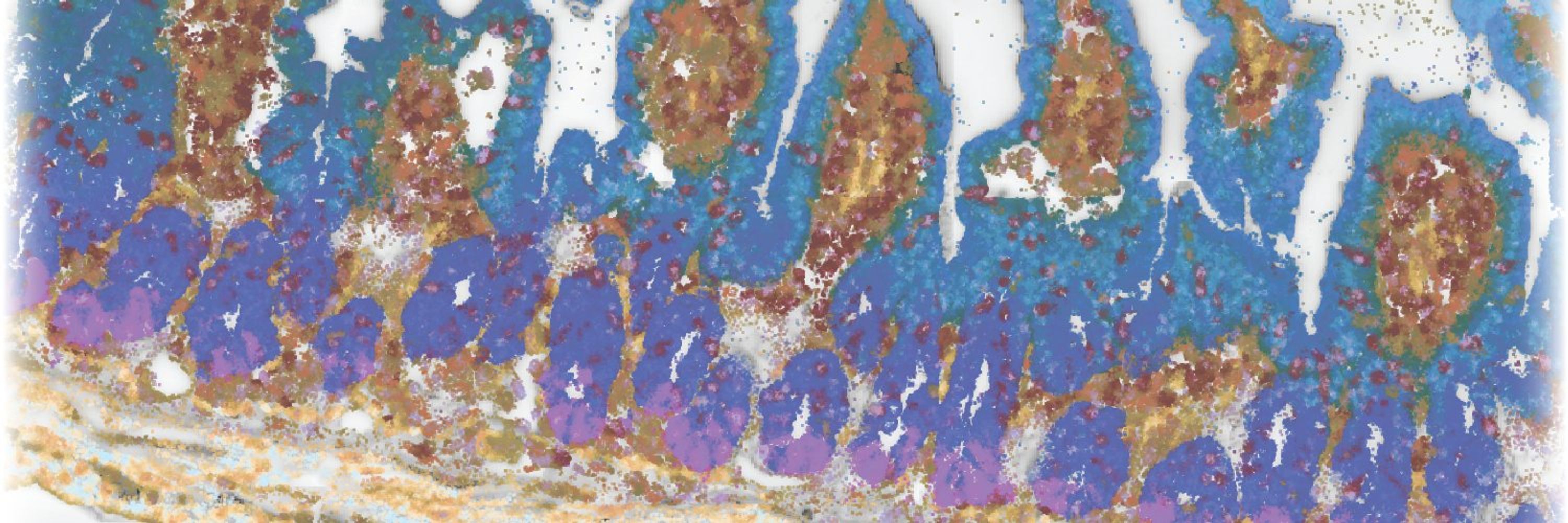
Excitingly, these advances enabled us to define a remarkable diversity of cell populations—78 in total—with finely resolved subdivisions within immune cells, fibroblasts, and the enteric nervous system! 4/
July 31, 2025 at 7:22 PM
Excitingly, these advances enabled us to define a remarkable diversity of cell populations—78 in total—with finely resolved subdivisions within immune cells, fibroblasts, and the enteric nervous system! 4/
As small molecule receptors are often lowly expressed even when functional, we developed new approaches to improve the sensitivity of our MERFISH measurements, including an improved cell segmentation pipeline. 3/
July 31, 2025 at 7:21 PM
As small molecule receptors are often lowly expressed even when functional, we developed new approaches to improve the sensitivity of our MERFISH measurements, including an improved cell segmentation pipeline. 3/
Excited to share our new work from Rosalind Xu, where we used MERFISH to build a single-cell spatial transcriptomic atlas of the mouse gut—and to explore host-microbiome interactions in situ! www.biorxiv.org/content/10.1... 1/
July 31, 2025 at 7:21 PM
Excited to share our new work from Rosalind Xu, where we used MERFISH to build a single-cell spatial transcriptomic atlas of the mouse gut—and to explore host-microbiome interactions in situ! www.biorxiv.org/content/10.1... 1/
Proud to announce our new technique for tracking relative B cell clonality in situ with B Cell Receptor MERFISH (BCR-MERFISH)! Congratulations to Evan Yang and our colleagues in the Carroll laboratory! Check out our bioRxiv: www.biorxiv.org/content/10.1... 1/9
May 28, 2025 at 3:40 PM
Proud to announce our new technique for tracking relative B cell clonality in situ with B Cell Receptor MERFISH (BCR-MERFISH)! Congratulations to Evan Yang and our colleagues in the Carroll laboratory! Check out our bioRxiv: www.biorxiv.org/content/10.1... 1/9
Remarkably, B. theta adapts its gene expression to different niches in the colon. For example, when near the host mucus layer, B. theta upregulates a rich diversity of genes associated with the harvest of mucus polysaccharides (PULs)! 12/
February 2, 2025 at 3:20 PM
Remarkably, B. theta adapts its gene expression to different niches in the colon. For example, when near the host mucus layer, B. theta upregulates a rich diversity of genes associated with the harvest of mucus polysaccharides (PULs)! 12/
The results were a shock! The E. coli transcriptome is extensively organized. We identified and classified a diversity of patterns and found that elements of the proteome organization and the genome organization shape the organization of the transcriptome. 10/
February 2, 2025 at 3:19 PM
The results were a shock! The E. coli transcriptome is extensively organized. We identified and classified a diversity of patterns and found that elements of the proteome organization and the genome organization shape the organization of the transcriptome. 10/
This heterogeneous response taught us two things. First, the required levels of essential genes during growth arises from coordinated transcriptional bursts. Second, when starved, E. coli explores many different sugars--not in our medium--before settling on xylose. 8/
February 2, 2025 at 3:18 PM
This heterogeneous response taught us two things. First, the required levels of essential genes during growth arises from coordinated transcriptional bursts. Second, when starved, E. coli explores many different sugars--not in our medium--before settling on xylose. 8/
We next revisited a classic experiment. E. coli grown in glucose and xylose will first consume glucose, then pause growth, before growing again on xylose. Classically, this pause is the time to make xylose-associated genes, but we observed a richer single-cell response. 7/
February 2, 2025 at 3:18 PM
We next revisited a classic experiment. E. coli grown in glucose and xylose will first consume glucose, then pause growth, before growing again on xylose. Classically, this pause is the time to make xylose-associated genes, but we observed a richer single-cell response. 7/

