Moffitt Lab
@moffittlab.bsky.social
Academic research lab passionate about building the next generation of genomic microscopy methods to reveal unknown features of the interface between microbe and host |
@bostonchildrens.bsky.social & @harvardmed.bsky.social | moffittlab.github.io
@bostonchildrens.bsky.social & @harvardmed.bsky.social | moffittlab.github.io
Remarkably, many aspects of the gut were largely unchanged! Nonetheless, we observed profound shifts in immune cell abundance and distribution—highlighting the central role of the microbiota in shaping and educating the immune system. 11/

July 31, 2025 at 7:24 PM
Remarkably, many aspects of the gut were largely unchanged! Nonetheless, we observed profound shifts in immune cell abundance and distribution—highlighting the central role of the microbiota in shaping and educating the immune system. 11/
The gut microbiota produces metabolites that impact host immunity and even CNS function—but how? Using a near-comprehensive receptor panel, we mapped metabolite sensitivity across gut cell types, revealing rich hypotheses for cell mediated microbiota–host interactions! 9/

July 31, 2025 at 7:23 PM
The gut microbiota produces metabolites that impact host immunity and even CNS function—but how? Using a near-comprehensive receptor panel, we mapped metabolite sensitivity across gut cell types, revealing rich hypotheses for cell mediated microbiota–host interactions! 9/
We also found a surprising set of genes with spatially patchy expression! In a great collaboration with Phillip Nicol (Irizarry lab), we developed a method to detect these patches —revealing insights into interferon signaling and epithelial specialization near GALT! 8/

July 31, 2025 at 7:23 PM
We also found a surprising set of genes with spatially patchy expression! In a great collaboration with Phillip Nicol (Irizarry lab), we developed a method to detect these patches —revealing insights into interferon signaling and epithelial specialization near GALT! 8/
This mucosal ruler revealed that nearly all cell populations fine-tune their gene expression based on their micron-scale position in the mucosa. Importantly, once one knows what to look for, these spatial signatures are also detectable in single-cell RNA-seq data! 7/

July 31, 2025 at 7:23 PM
This mucosal ruler revealed that nearly all cell populations fine-tune their gene expression based on their micron-scale position in the mucosa. Importantly, once one knows what to look for, these spatial signatures are also detectable in single-cell RNA-seq data! 7/
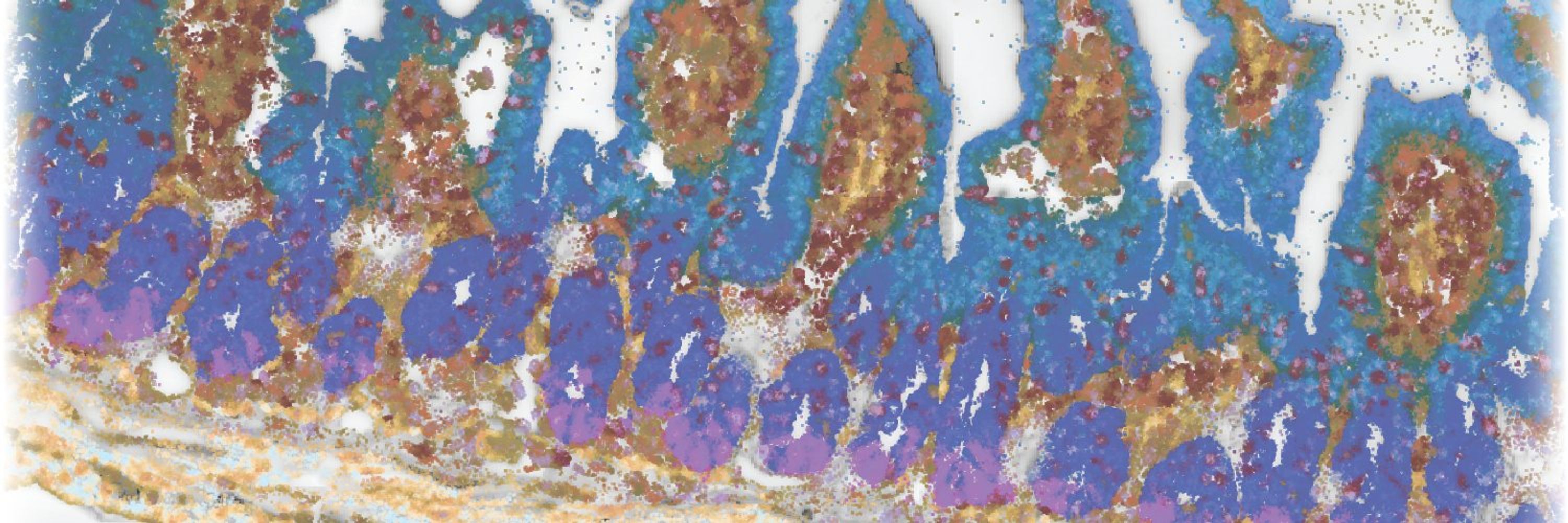
Another surprise was the intricate spatial organization of cells and RNAs in the mucosa. From crypt base to lumen, we observed gradients in cell types and gene expression, some known, many novel! We created an expression-based mucosal spatial ruler to capture this pattern. 6/

July 31, 2025 at 7:22 PM
Another surprise was the intricate spatial organization of cells and RNAs in the mucosa. From crypt base to lumen, we observed gradients in cell types and gene expression, some known, many novel! We created an expression-based mucosal spatial ruler to capture this pattern. 6/
Our atlas revealed multiple surprises! For instance, we found a novel mature enterocyte marked by Scnn1g, a subunit of the epithelial sodium channel, and the pseudogene Best4-ps —suggesting it might be the long-sought mouse homolog of the human BEST4+ enterocyte! 5/

July 31, 2025 at 7:22 PM
Our atlas revealed multiple surprises! For instance, we found a novel mature enterocyte marked by Scnn1g, a subunit of the epithelial sodium channel, and the pseudogene Best4-ps —suggesting it might be the long-sought mouse homolog of the human BEST4+ enterocyte! 5/
Excitingly, these advances enabled us to define a remarkable diversity of cell populations—78 in total—with finely resolved subdivisions within immune cells, fibroblasts, and the enteric nervous system! 4/
July 31, 2025 at 7:22 PM
Excitingly, these advances enabled us to define a remarkable diversity of cell populations—78 in total—with finely resolved subdivisions within immune cells, fibroblasts, and the enteric nervous system! 4/
As small molecule receptors are often lowly expressed even when functional, we developed new approaches to improve the sensitivity of our MERFISH measurements, including an improved cell segmentation pipeline. 3/
July 31, 2025 at 7:21 PM
As small molecule receptors are often lowly expressed even when functional, we developed new approaches to improve the sensitivity of our MERFISH measurements, including an improved cell segmentation pipeline. 3/
Excited to share our new work from Rosalind Xu, where we used MERFISH to build a single-cell spatial transcriptomic atlas of the mouse gut—and to explore host-microbiome interactions in situ! www.biorxiv.org/content/10.1... 1/
July 31, 2025 at 7:21 PM
Excited to share our new work from Rosalind Xu, where we used MERFISH to build a single-cell spatial transcriptomic atlas of the mouse gut—and to explore host-microbiome interactions in situ! www.biorxiv.org/content/10.1... 1/
As small molecule receptors are often lowly expressed even when functional, we developed new approaches to improve the sensitivity of our MERFISH measurements, including an improved cell segmentation pipeline. 3/

July 31, 2025 at 7:20 PM
As small molecule receptors are often lowly expressed even when functional, we developed new approaches to improve the sensitivity of our MERFISH measurements, including an improved cell segmentation pipeline. 3/
Yesterday our first grad student -- DR. Rosalind Xu -- had her official PhD hooding ceremony in Harvard CCB! A massive congratulations to her and all of the other freshly minted PhDs!

May 30, 2025 at 12:38 PM
Yesterday our first grad student -- DR. Rosalind Xu -- had her official PhD hooding ceremony in Harvard CCB! A massive congratulations to her and all of the other freshly minted PhDs!
To our surprise, we found that some plasma cell clones form patches of co-occurring clones along the gut, suggesting, perhaps, that there is local production of IgA specific to a subset of the microbiome! 8/9

May 28, 2025 at 3:43 PM
To our surprise, we found that some plasma cell clones form patches of co-occurring clones along the gut, suggesting, perhaps, that there is local production of IgA specific to a subset of the microbiome! 8/9
Plasma cells modulate the microbiome by secreting IgA, and different plasma cell clones secrete IgA that targets different bacteria. But how these cells are distributed along the gut is not clear. Excitingly, BCR-MERFISH can now measure this distribution. 7/9

May 28, 2025 at 3:42 PM
Plasma cells modulate the microbiome by secreting IgA, and different plasma cell clones secrete IgA that targets different bacteria. But how these cells are distributed along the gut is not clear. Excitingly, BCR-MERFISH can now measure this distribution. 7/9
Importantly, BCR-MERFISH labels not only the Clone ID, but also the transcriptome of all surrounding cell types in the same slice, placing unique clones within their tissue contexts with single-cell resolution. 6/9

May 28, 2025 at 3:42 PM
Importantly, BCR-MERFISH labels not only the Clone ID, but also the transcriptome of all surrounding cell types in the same slice, placing unique clones within their tissue contexts with single-cell resolution. 6/9
To test BCR-MERFISH, we profiled V gene usage in plasma B cells in the mouse ileum, defined B cell clones via the VH and VKVL gene choice within individual B cells, and showed that the usage of these genes agreed with BCR-sequencing. 5/9

May 28, 2025 at 3:42 PM
To test BCR-MERFISH, we profiled V gene usage in plasma B cells in the mouse ileum, defined B cell clones via the VH and VKVL gene choice within individual B cells, and showed that the usage of these genes agreed with BCR-sequencing. 5/9
However, MERFISH has been unable to discriminate B cells based on the BCR sequence because of the high degree of homology between V, D, and J genes. Evan solved this problem with a homology-aware probe design and encoding approach to produce a technique we call BCR-MERFISH! 4/9

May 28, 2025 at 3:41 PM
However, MERFISH has been unable to discriminate B cells based on the BCR sequence because of the high degree of homology between V, D, and J genes. Evan solved this problem with a homology-aware probe design and encoding approach to produce a technique we call BCR-MERFISH! 4/9
Proud to announce our new technique for tracking relative B cell clonality in situ with B Cell Receptor MERFISH (BCR-MERFISH)! Congratulations to Evan Yang and our colleagues in the Carroll laboratory! Check out our bioRxiv: www.biorxiv.org/content/10.1... 1/9
May 28, 2025 at 3:40 PM
Proud to announce our new technique for tracking relative B cell clonality in situ with B Cell Receptor MERFISH (BCR-MERFISH)! Congratulations to Evan Yang and our colleagues in the Carroll laboratory! Check out our bioRxiv: www.biorxiv.org/content/10.1... 1/9
Remarkably, B. theta adapts its gene expression to different niches in the colon. For example, when near the host mucus layer, B. theta upregulates a rich diversity of genes associated with the harvest of mucus polysaccharides (PULs)! 12/
February 2, 2025 at 3:20 PM
Remarkably, B. theta adapts its gene expression to different niches in the colon. For example, when near the host mucus layer, B. theta upregulates a rich diversity of genes associated with the harvest of mucus polysaccharides (PULs)! 12/
Most bacteria live in complex, spatially structured environments not well approximated by a culture flask. To illustrate the ability of bacterial-MERFISH to profile bacterial gene expression in such environments, we characterized B. theta expression in the colon of mice. 11/

February 2, 2025 at 3:20 PM
Most bacteria live in complex, spatially structured environments not well approximated by a culture flask. To illustrate the ability of bacterial-MERFISH to profile bacterial gene expression in such environments, we characterized B. theta expression in the colon of mice. 11/
The results were a shock! The E. coli transcriptome is extensively organized. We identified and classified a diversity of patterns and found that elements of the proteome organization and the genome organization shape the organization of the transcriptome. 10/
February 2, 2025 at 3:19 PM
The results were a shock! The E. coli transcriptome is extensively organized. We identified and classified a diversity of patterns and found that elements of the proteome organization and the genome organization shape the organization of the transcriptome. 10/
Even though bacteria are small, there is a growing appreciation that different mRNAs can be localized to different sub-cellular regions with important functional consequences. So next we charted the organization of the E. coli transcriptome with bacterial-MERFISH. 9/

February 2, 2025 at 3:19 PM
Even though bacteria are small, there is a growing appreciation that different mRNAs can be localized to different sub-cellular regions with important functional consequences. So next we charted the organization of the E. coli transcriptome with bacterial-MERFISH. 9/
This heterogeneous response taught us two things. First, the required levels of essential genes during growth arises from coordinated transcriptional bursts. Second, when starved, E. coli explores many different sugars--not in our medium--before settling on xylose. 8/
February 2, 2025 at 3:18 PM
This heterogeneous response taught us two things. First, the required levels of essential genes during growth arises from coordinated transcriptional bursts. Second, when starved, E. coli explores many different sugars--not in our medium--before settling on xylose. 8/
We next revisited a classic experiment. E. coli grown in glucose and xylose will first consume glucose, then pause growth, before growing again on xylose. Classically, this pause is the time to make xylose-associated genes, but we observed a richer single-cell response. 7/
February 2, 2025 at 3:18 PM
We next revisited a classic experiment. E. coli grown in glucose and xylose will first consume glucose, then pause growth, before growing again on xylose. Classically, this pause is the time to make xylose-associated genes, but we observed a richer single-cell response. 7/
Expanded bacteria are then combined with #MERFISH to image large fractions of the transcriptome. Benchmarked in E. coli, bacterial-MERFISH is accurate across a large dynamic range, has high detection efficiency and low false positives, and can image 100k+ cells. 6/

February 2, 2025 at 3:17 PM
Expanded bacteria are then combined with #MERFISH to image large fractions of the transcriptome. Benchmarked in E. coli, bacterial-MERFISH is accurate across a large dynamic range, has high detection efficiency and low false positives, and can image 100k+ cells. 6/
However, three orders of magnitude didn’t seem so daunting to Ari, Yuanyou, and Nana, so they built a bacteria-optimized #ExpansionMicroscopy toolbox that is capable of tunable expansion of individual bacteria up to 1000-fold in volume! 5/

February 2, 2025 at 3:16 PM
However, three orders of magnitude didn’t seem so daunting to Ari, Yuanyou, and Nana, so they built a bacteria-optimized #ExpansionMicroscopy toolbox that is capable of tunable expansion of individual bacteria up to 1000-fold in volume! 5/

