Miro Astore
@miroastore.bsky.social
Postdoc @rockefeller. Making cryoEM into a tool for RNA biophysics one molecules at a time.
The text of the order says it's a one time charge for new petitions, renewals and transfers.
www.whitehouse.gov/presidential...
www.whitehouse.gov/presidential...

Restriction on Entry of Certain Nonimmigrant Workers
BY THE PRESIDENT OF THE UNITED STATES OF AMERICA A PROCLAMATION The H-1B nonimmigrant visa program was created to bring temporary workers into the United
www.whitehouse.gov
September 20, 2025 at 10:49 PM
The text of the order says it's a one time charge for new petitions, renewals and transfers.
www.whitehouse.gov/presidential...
www.whitehouse.gov/presidential...
Link that works
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

The Inaugural Flatiron Institute Cryo-EM Conformational Heterogeneity Challenge
Despite the rise of single particle cryo-electron microscopy (cryo-EM) as a premier method for resolving macromolecular structures at atomic resolution, methods to address molecular heterogeneity in v...
www.biorxiv.org
July 24, 2025 at 10:43 AM
Link that works
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
You can also check out the analysis workflow and the datasets themselves if you'd like to learn more github.com/flatironinst...
www.simonsfoundation.org/flatiron/cen...
www.simonsfoundation.org/flatiron/cen...

GitHub - flatironinstitute/Cryo-EM-Heterogeneity-Challenge-1: The Inaugural Flatiron Institute Cryo-EM Heterogeneity Community Challenge
The Inaugural Flatiron Institute Cryo-EM Heterogeneity Community Challenge - flatironinstitute/Cryo-EM-Heterogeneity-Challenge-1
github.com
July 23, 2025 at 9:52 PM
You can also check out the analysis workflow and the datasets themselves if you'd like to learn more github.com/flatironinst...
www.simonsfoundation.org/flatiron/cen...
www.simonsfoundation.org/flatiron/cen...
This has been so much work from my coauthors Geoff Woollard and David Silva-Sanchez, mentors @sonyahanson.bsky.social
and @pilarcossio.bsky.social , Misha Kopylov at NYSBC, and all the participants who have continuously given us excellent feedback on the study and the manuscript. Thanks everybody!
and @pilarcossio.bsky.social , Misha Kopylov at NYSBC, and all the participants who have continuously given us excellent feedback on the study and the manuscript. Thanks everybody!
July 23, 2025 at 9:43 PM
This has been so much work from my coauthors Geoff Woollard and David Silva-Sanchez, mentors @sonyahanson.bsky.social
and @pilarcossio.bsky.social , Misha Kopylov at NYSBC, and all the participants who have continuously given us excellent feedback on the study and the manuscript. Thanks everybody!
and @pilarcossio.bsky.social , Misha Kopylov at NYSBC, and all the participants who have continuously given us excellent feedback on the study and the manuscript. Thanks everybody!
The last metric we used calculates the likelihood that a given ensemble of volumes is contained in a set of images. This one is really exciting because we don't need any ground truth to compare to. We see the same trends between the simulated and experimental datasets.

July 23, 2025 at 9:43 PM
The last metric we used calculates the likelihood that a given ensemble of volumes is contained in a set of images. This one is really exciting because we don't need any ground truth to compare to. We see the same trends between the simulated and experimental datasets.
The next is based on optimal transport. By calculating the differences between maps in a submission and the ground truth, we can calculate how close a submission is to the truth. We can also see how far from optimal participants estimated their populations.

July 23, 2025 at 9:43 PM
The next is based on optimal transport. By calculating the differences between maps in a submission and the ground truth, we can calculate how close a submission is to the truth. We can also see how far from optimal participants estimated their populations.
Now comes the real work. Comparing all this amazing data! We came up with three novel ways of comparing series of volumes. With the first we simply perform PCA on the voxel maps and compare their bases. Turns out to be a super useful way of comparing motions between submissions.

July 23, 2025 at 9:43 PM
Now comes the real work. Comparing all this amazing data! We came up with three novel ways of comparing series of volumes. With the first we simply perform PCA on the voxel maps and compare their bases. Turns out to be a super useful way of comparing motions between submissions.
We had amazing participation. Almost every method in the literature submitted to this challenge. We've chosen to keep the identity of the submissions anonymous (you'll see them in the manuscript denoted by icecream flavor hehe).

July 23, 2025 at 9:43 PM
We had amazing participation. Almost every method in the literature submitted to this challenge. We've chosen to keep the identity of the submissions anonymous (you'll see them in the manuscript denoted by icecream flavor hehe).
We asked each participant to give us two things. 80 volume maps which they thought gave a good representation of the molecular states in the data. And relative populations between those molecular states.
July 23, 2025 at 9:43 PM
We asked each participant to give us two things. 80 volume maps which they thought gave a good representation of the molecular states in the data. And relative populations between those molecular states.
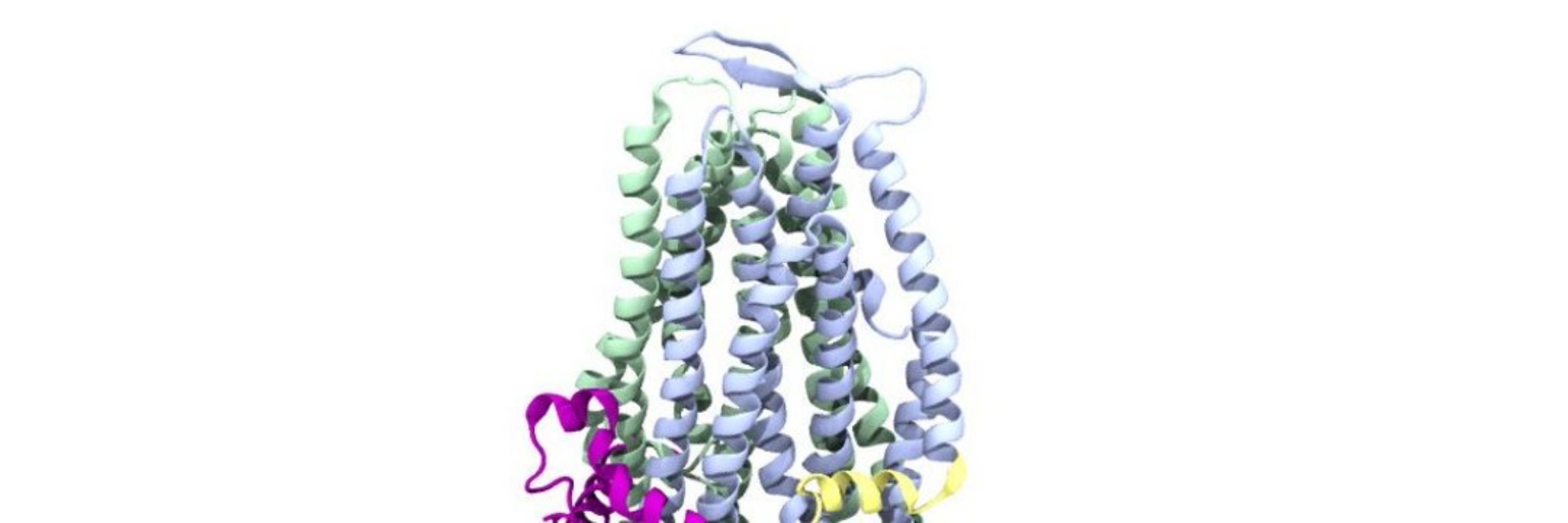
That's why we launched this challenge. We made two datasets, one experimental and one simulated. The simulated one was made with structures from molecular dynamics simulations. Here we hid a triple Gaussian for participants to discover. (Movie credit David Herreros).
July 23, 2025 at 9:43 PM
That's why we launched this challenge. We made two datasets, one experimental and one simulated. The simulated one was made with structures from molecular dynamics simulations. Here we hid a triple Gaussian for participants to discover. (Movie credit David Herreros).
There has been an explosion of methods to determine molecular ensembles from single particle cryo-EM datasets. However, these methods all use totally different algorithms and it's currently unknown how to assess their accuracy without ground truth structures to compare with.

July 23, 2025 at 9:43 PM
There has been an explosion of methods to determine molecular ensembles from single particle cryo-EM datasets. However, these methods all use totally different algorithms and it's currently unknown how to assess their accuracy without ground truth structures to compare with.

